| Full name: N-acetyltransferase 9 (putative) | Alias Symbol: DKFZP564C103 | ||
| Type: protein-coding gene | Cytoband: 17q25.2 | ||
| Entrez ID: 26151 | HGNC ID: HGNC:23133 | Ensembl Gene: ENSG00000109065 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of NAT9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAT9 | 26151 | 204382_at | 0.7496 | 0.1184 | |
| GSE20347 | NAT9 | 26151 | 204382_at | 0.7354 | 0.0000 | |
| GSE23400 | NAT9 | 26151 | 204382_at | 0.4695 | 0.0000 | |
| GSE26886 | NAT9 | 26151 | 204382_at | 0.4063 | 0.0179 | |
| GSE29001 | NAT9 | 26151 | 204382_at | 0.5950 | 0.0025 | |
| GSE38129 | NAT9 | 26151 | 204382_at | 0.5519 | 0.0000 | |
| GSE45670 | NAT9 | 26151 | 204382_at | 0.5133 | 0.0067 | |
| GSE53622 | NAT9 | 26151 | 54101 | 0.5065 | 0.0000 | |
| GSE53624 | NAT9 | 26151 | 54101 | 0.7229 | 0.0000 | |
| GSE63941 | NAT9 | 26151 | 204382_at | 0.2541 | 0.4321 | |
| GSE77861 | NAT9 | 26151 | 204382_at | 0.4669 | 0.0026 | |
| GSE97050 | NAT9 | 26151 | A_23_P66421 | -0.1048 | 0.6169 | |
| SRP007169 | NAT9 | 26151 | RNAseq | 1.2279 | 0.0148 | |
| SRP008496 | NAT9 | 26151 | RNAseq | 1.2906 | 0.0011 | |
| SRP064894 | NAT9 | 26151 | RNAseq | 0.9503 | 0.0000 | |
| SRP133303 | NAT9 | 26151 | RNAseq | 0.4686 | 0.0377 | |
| SRP159526 | NAT9 | 26151 | RNAseq | 0.7048 | 0.0232 | |
| SRP193095 | NAT9 | 26151 | RNAseq | 0.4541 | 0.0000 | |
| SRP219564 | NAT9 | 26151 | RNAseq | 1.2892 | 0.0325 | |
| TCGA | NAT9 | 26151 | RNAseq | 0.1026 | 0.1241 |
Upregulated datasets: 3; Downregulated datasets: 0.
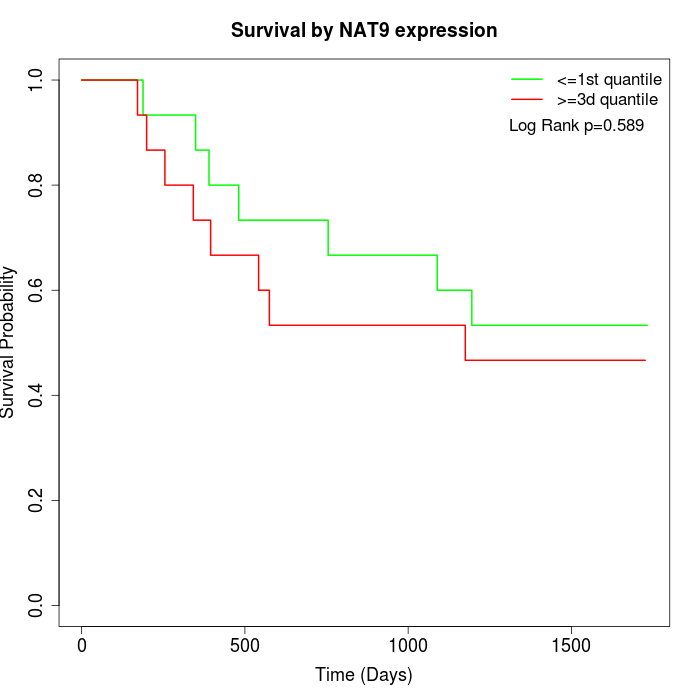
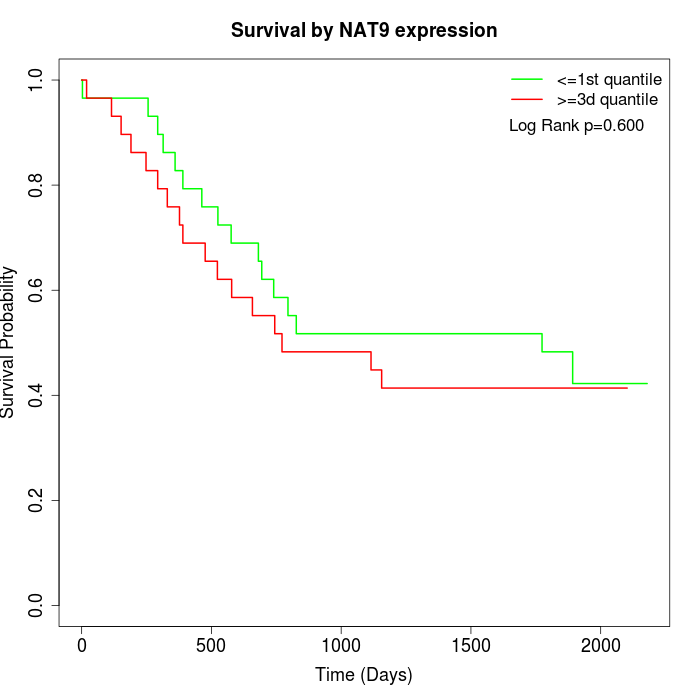
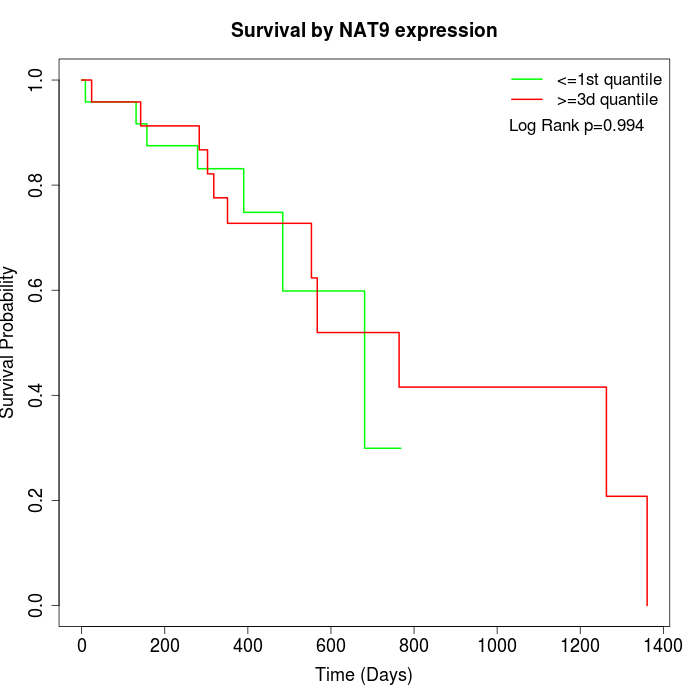
Survival by NAT9 expression:
Note: Click image to view full size file.
Copy number change of NAT9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAT9 | 26151 | 3 | 2 | 25 | |
| GSE20123 | NAT9 | 26151 | 3 | 2 | 25 | |
| GSE43470 | NAT9 | 26151 | 5 | 3 | 35 | |
| GSE46452 | NAT9 | 26151 | 33 | 0 | 26 | |
| GSE47630 | NAT9 | 26151 | 8 | 1 | 31 | |
| GSE54993 | NAT9 | 26151 | 2 | 5 | 63 | |
| GSE54994 | NAT9 | 26151 | 10 | 4 | 39 | |
| GSE60625 | NAT9 | 26151 | 6 | 0 | 5 | |
| GSE74703 | NAT9 | 26151 | 5 | 1 | 30 | |
| GSE74704 | NAT9 | 26151 | 3 | 1 | 16 | |
| TCGA | NAT9 | 26151 | 32 | 10 | 54 |
Total number of gains: 110; Total number of losses: 29; Total Number of normals: 349.
Somatic mutations of NAT9:
Generating mutation plots.
Highly correlated genes for NAT9:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAT9 | TIGD7 | 0.720747 | 3 | 0 | 3 |
| NAT9 | IPO9 | 0.700425 | 7 | 0 | 7 |
| NAT9 | C19orf25 | 0.69909 | 4 | 0 | 3 |
| NAT9 | PAG1 | 0.698998 | 3 | 0 | 3 |
| NAT9 | ARL8A | 0.697833 | 3 | 0 | 3 |
| NAT9 | CADM4 | 0.694719 | 3 | 0 | 3 |
| NAT9 | CPAMD8 | 0.686282 | 3 | 0 | 3 |
| NAT9 | MAF1 | 0.681325 | 3 | 0 | 3 |
| NAT9 | C20orf144 | 0.67182 | 3 | 0 | 3 |
| NAT9 | FBRSL1 | 0.667062 | 5 | 0 | 4 |
| NAT9 | RBM28 | 0.664927 | 11 | 0 | 9 |
| NAT9 | RFX8 | 0.662994 | 3 | 0 | 3 |
| NAT9 | UBALD2 | 0.658364 | 3 | 0 | 3 |
| NAT9 | ZNF696 | 0.654363 | 9 | 0 | 9 |
| NAT9 | NAT14 | 0.650419 | 6 | 0 | 5 |
| NAT9 | CCT3 | 0.6484 | 9 | 0 | 7 |
| NAT9 | SOGA1 | 0.647472 | 4 | 0 | 3 |
| NAT9 | KLK3 | 0.645124 | 3 | 0 | 3 |
| NAT9 | LRRC71 | 0.644357 | 3 | 0 | 3 |
| NAT9 | POLR2H | 0.642254 | 10 | 0 | 8 |
For details and further investigation, click here