| Full name: calcium voltage-gated channel auxiliary subunit beta 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10p12 | ||
| Entrez ID: 783 | HGNC ID: HGNC:1402 | Ensembl Gene: ENSG00000165995 | OMIM ID: 600003 |
| Drug and gene relationship at DGIdb | |||
CACNB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa04921 | Oxytocin signaling pathway |
Expression of CACNB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CACNB2 | 783 | 207776_s_at | -1.5103 | 0.1313 | |
| GSE20347 | CACNB2 | 783 | 207776_s_at | -0.1073 | 0.3681 | |
| GSE23400 | CACNB2 | 783 | 207776_s_at | -0.2074 | 0.0287 | |
| GSE26886 | CACNB2 | 783 | 1555098_a_at | 0.1539 | 0.1526 | |
| GSE29001 | CACNB2 | 783 | 207776_s_at | -0.0825 | 0.6803 | |
| GSE38129 | CACNB2 | 783 | 207776_s_at | -0.4913 | 0.0008 | |
| GSE45670 | CACNB2 | 783 | 207776_s_at | -1.2155 | 0.0000 | |
| GSE53622 | CACNB2 | 783 | 30445 | -2.0068 | 0.0000 | |
| GSE53624 | CACNB2 | 783 | 30445 | -1.3188 | 0.0000 | |
| GSE63941 | CACNB2 | 783 | 207776_s_at | -0.2295 | 0.3603 | |
| GSE77861 | CACNB2 | 783 | 215365_at | -0.0889 | 0.4332 | |
| GSE97050 | CACNB2 | 783 | A_33_P3285277 | -1.4282 | 0.0966 | |
| SRP007169 | CACNB2 | 783 | RNAseq | 2.2530 | 0.0116 | |
| SRP064894 | CACNB2 | 783 | RNAseq | -0.4648 | 0.1696 | |
| SRP133303 | CACNB2 | 783 | RNAseq | -0.5347 | 0.1843 | |
| SRP159526 | CACNB2 | 783 | RNAseq | -0.4911 | 0.3302 | |
| SRP193095 | CACNB2 | 783 | RNAseq | -0.0932 | 0.5350 | |
| SRP219564 | CACNB2 | 783 | RNAseq | -0.7799 | 0.3838 | |
| TCGA | CACNB2 | 783 | RNAseq | -1.0144 | 0.0001 |
Upregulated datasets: 1; Downregulated datasets: 4.
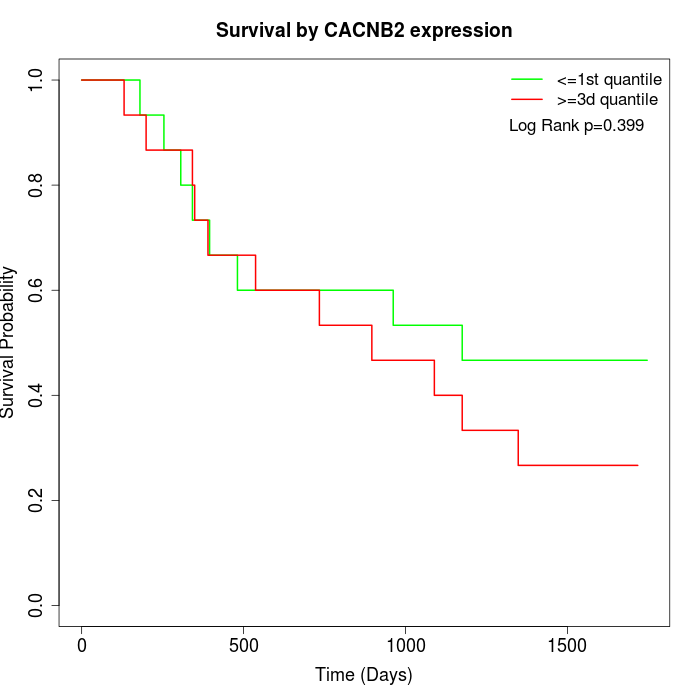
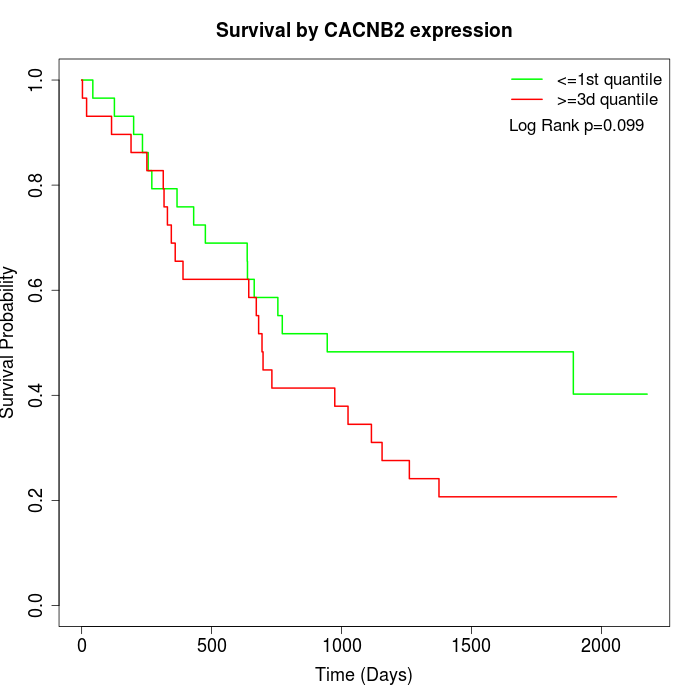
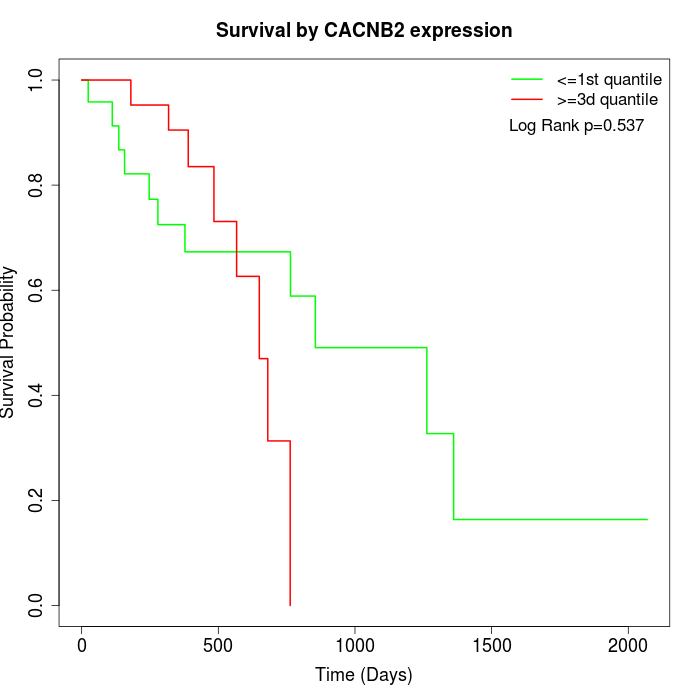
Survival by CACNB2 expression:
Note: Click image to view full size file.
Copy number change of CACNB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CACNB2 | 783 | 3 | 8 | 19 | |
| GSE20123 | CACNB2 | 783 | 3 | 7 | 20 | |
| GSE43470 | CACNB2 | 783 | 3 | 4 | 36 | |
| GSE46452 | CACNB2 | 783 | 1 | 14 | 44 | |
| GSE47630 | CACNB2 | 783 | 5 | 15 | 20 | |
| GSE54993 | CACNB2 | 783 | 10 | 0 | 60 | |
| GSE54994 | CACNB2 | 783 | 4 | 9 | 40 | |
| GSE60625 | CACNB2 | 783 | 0 | 0 | 11 | |
| GSE74703 | CACNB2 | 783 | 2 | 3 | 31 | |
| GSE74704 | CACNB2 | 783 | 0 | 6 | 14 | |
| TCGA | CACNB2 | 783 | 18 | 26 | 52 |
Total number of gains: 49; Total number of losses: 92; Total Number of normals: 347.
Somatic mutations of CACNB2:
Generating mutation plots.
Highly correlated genes for CACNB2:
Showing top 20/1036 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CACNB2 | MYOCD | 0.922918 | 5 | 0 | 5 |
| CACNB2 | ANGPTL1 | 0.890844 | 5 | 0 | 5 |
| CACNB2 | SORCS1 | 0.873892 | 4 | 0 | 4 |
| CACNB2 | XKR4 | 0.851051 | 5 | 0 | 5 |
| CACNB2 | NEGR1 | 0.847334 | 5 | 0 | 5 |
| CACNB2 | ANO5 | 0.845549 | 5 | 0 | 5 |
| CACNB2 | MRVI1 | 0.838741 | 6 | 0 | 6 |
| CACNB2 | MAMDC2 | 0.825579 | 5 | 0 | 5 |
| CACNB2 | MIR100HG | 0.824926 | 4 | 0 | 4 |
| CACNB2 | PPP1R12B | 0.824063 | 8 | 0 | 8 |
| CACNB2 | C1QTNF7 | 0.814628 | 5 | 0 | 5 |
| CACNB2 | MSRB3 | 0.811925 | 5 | 0 | 5 |
| CACNB2 | CLEC3B | 0.808224 | 3 | 0 | 3 |
| CACNB2 | ITGA9 | 0.805349 | 6 | 0 | 6 |
| CACNB2 | BHMT2 | 0.804356 | 8 | 0 | 8 |
| CACNB2 | PTCHD1 | 0.798535 | 5 | 0 | 5 |
| CACNB2 | ATP1A2 | 0.791701 | 8 | 0 | 7 |
| CACNB2 | ASB5 | 0.791517 | 6 | 0 | 5 |
| CACNB2 | PDZRN4 | 0.790296 | 8 | 0 | 7 |
| CACNB2 | VMAC | 0.789407 | 3 | 0 | 3 |
For details and further investigation, click here