| Full name: NPHS2 stomatin family member, podocin | Alias Symbol: SRN1|PDCN | ||
| Type: protein-coding gene | Cytoband: 1q25.2 | ||
| Entrez ID: 7827 | HGNC ID: HGNC:13394 | Ensembl Gene: ENSG00000116218 | OMIM ID: 604766 |
| Drug and gene relationship at DGIdb | |||
Expression of NPHS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NPHS2 | 7827 | 220424_at | -0.0440 | 0.8697 | |
| GSE20347 | NPHS2 | 7827 | 220424_at | -0.0637 | 0.5748 | |
| GSE23400 | NPHS2 | 7827 | 220424_at | -0.2311 | 0.0000 | |
| GSE26886 | NPHS2 | 7827 | 220424_at | -0.2807 | 0.0651 | |
| GSE29001 | NPHS2 | 7827 | 220424_at | -0.2947 | 0.2254 | |
| GSE38129 | NPHS2 | 7827 | 220424_at | -0.1546 | 0.1233 | |
| GSE45670 | NPHS2 | 7827 | 220424_at | 0.0781 | 0.5258 | |
| GSE53622 | NPHS2 | 7827 | 11306 | -0.1010 | 0.2843 | |
| GSE53624 | NPHS2 | 7827 | 11306 | 0.3610 | 0.0304 | |
| GSE63941 | NPHS2 | 7827 | 220424_at | -0.0497 | 0.7957 | |
| GSE77861 | NPHS2 | 7827 | 220424_at | -0.2273 | 0.0986 | |
| TCGA | NPHS2 | 7827 | RNAseq | -1.5245 | 0.2485 |
Upregulated datasets: 0; Downregulated datasets: 0.
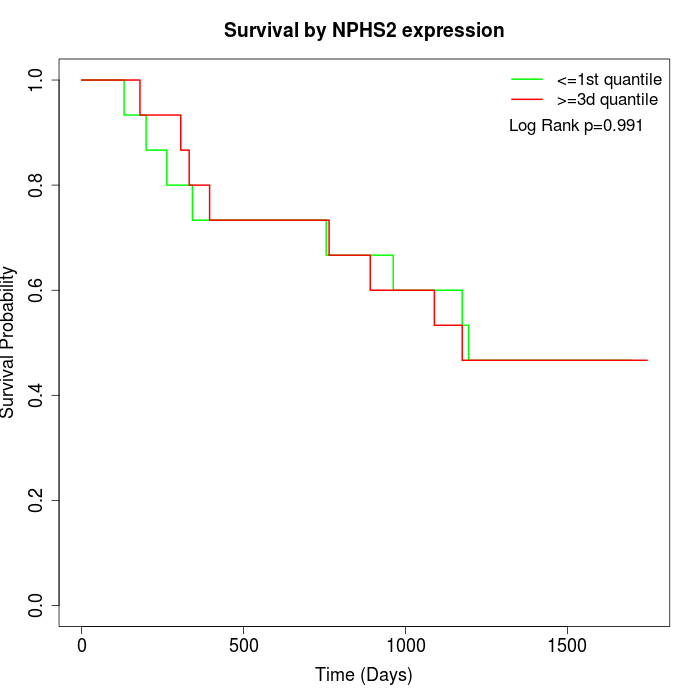
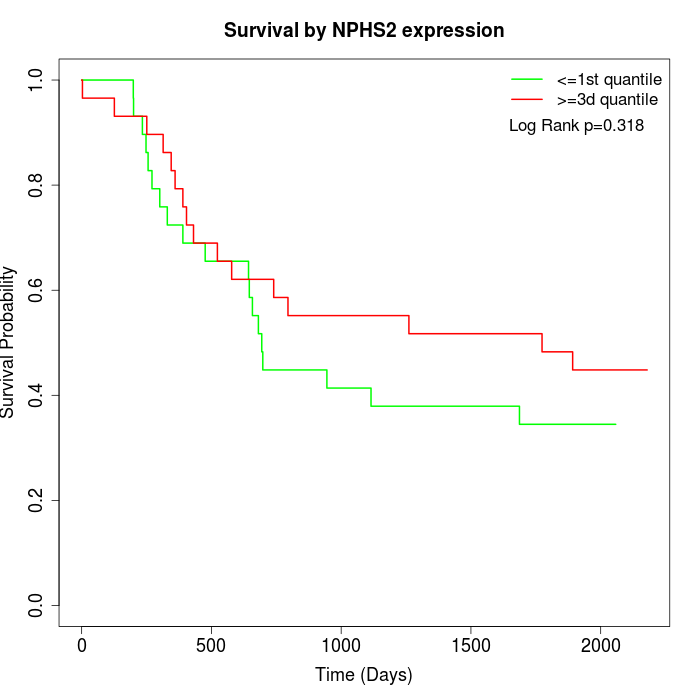
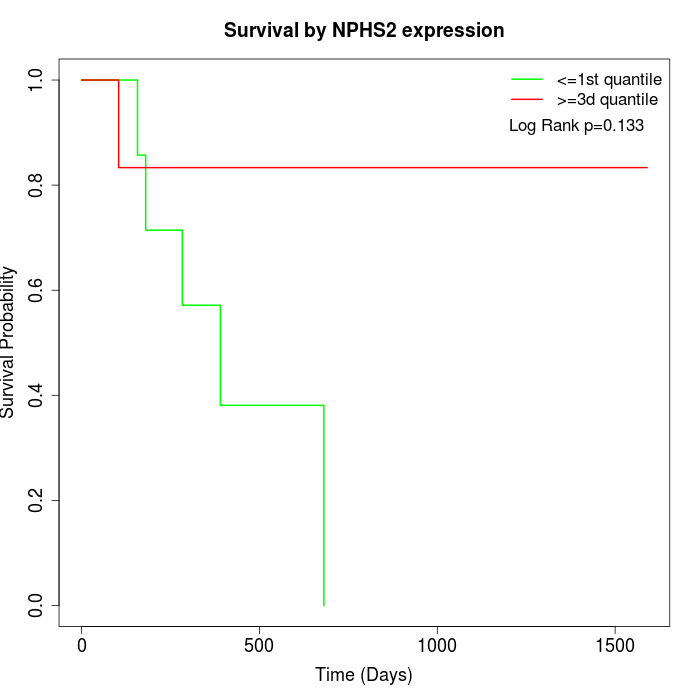
Survival by NPHS2 expression:
Note: Click image to view full size file.
Copy number change of NPHS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NPHS2 | 7827 | 12 | 0 | 18 | |
| GSE20123 | NPHS2 | 7827 | 12 | 0 | 18 | |
| GSE43470 | NPHS2 | 7827 | 7 | 1 | 35 | |
| GSE46452 | NPHS2 | 7827 | 3 | 1 | 55 | |
| GSE47630 | NPHS2 | 7827 | 14 | 0 | 26 | |
| GSE54993 | NPHS2 | 7827 | 0 | 6 | 64 | |
| GSE54994 | NPHS2 | 7827 | 15 | 0 | 38 | |
| GSE60625 | NPHS2 | 7827 | 0 | 0 | 11 | |
| GSE74703 | NPHS2 | 7827 | 7 | 1 | 28 | |
| GSE74704 | NPHS2 | 7827 | 5 | 0 | 15 | |
| TCGA | NPHS2 | 7827 | 40 | 3 | 53 |
Total number of gains: 115; Total number of losses: 12; Total Number of normals: 361.
Somatic mutations of NPHS2:
Generating mutation plots.
Highly correlated genes for NPHS2:
Showing top 20/957 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NPHS2 | USP2 | 0.749794 | 4 | 0 | 4 |
| NPHS2 | CA5A | 0.745134 | 5 | 0 | 5 |
| NPHS2 | INO80D | 0.744044 | 4 | 0 | 4 |
| NPHS2 | CYP19A1 | 0.727522 | 4 | 0 | 4 |
| NPHS2 | VWA7 | 0.722418 | 5 | 0 | 5 |
| NPHS2 | STARD13-AS | 0.718726 | 3 | 0 | 3 |
| NPHS2 | CYP2D6 | 0.717806 | 5 | 0 | 5 |
| NPHS2 | NTNG1 | 0.707805 | 4 | 0 | 4 |
| NPHS2 | MTAP | 0.706382 | 4 | 0 | 4 |
| NPHS2 | JAK3 | 0.706098 | 4 | 0 | 3 |
| NPHS2 | IL1RL1 | 0.705705 | 4 | 0 | 4 |
| NPHS2 | E2F4 | 0.705237 | 7 | 0 | 7 |
| NPHS2 | XPNPEP2 | 0.703858 | 3 | 0 | 3 |
| NPHS2 | IRF2BP1 | 0.703091 | 3 | 0 | 3 |
| NPHS2 | C11orf16 | 0.702321 | 5 | 0 | 5 |
| NPHS2 | SHBG | 0.69997 | 6 | 0 | 5 |
| NPHS2 | EPOR | 0.699842 | 8 | 0 | 7 |
| NPHS2 | CBL | 0.698664 | 4 | 0 | 4 |
| NPHS2 | ADAM22 | 0.695511 | 5 | 0 | 4 |
| NPHS2 | SERPINA4 | 0.694698 | 5 | 0 | 5 |
For details and further investigation, click here