| Full name: carbonic anhydrase 5A | Alias Symbol: CAV|CAVA | ||
| Type: protein-coding gene | Cytoband: 16q24.2 | ||
| Entrez ID: 763 | HGNC ID: HGNC:1377 | Ensembl Gene: ENSG00000174990 | OMIM ID: 114761 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CA5A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CA5A | 763 | 207421_at | -0.0981 | 0.6730 | |
| GSE20347 | CA5A | 763 | 207421_at | 0.0014 | 0.9885 | |
| GSE23400 | CA5A | 763 | 207421_at | -0.1832 | 0.0000 | |
| GSE26886 | CA5A | 763 | 207421_at | 0.1286 | 0.3492 | |
| GSE29001 | CA5A | 763 | 207421_at | -0.1757 | 0.2292 | |
| GSE38129 | CA5A | 763 | 207421_at | -0.0624 | 0.3617 | |
| GSE45670 | CA5A | 763 | 207421_at | 0.0977 | 0.2704 | |
| GSE63941 | CA5A | 763 | 207421_at | 0.2277 | 0.0481 | |
| GSE77861 | CA5A | 763 | 207421_at | -0.0300 | 0.8094 | |
| GSE97050 | CA5A | 763 | A_24_P540057 | -0.6885 | 0.2136 |
Upregulated datasets: 0; Downregulated datasets: 0.
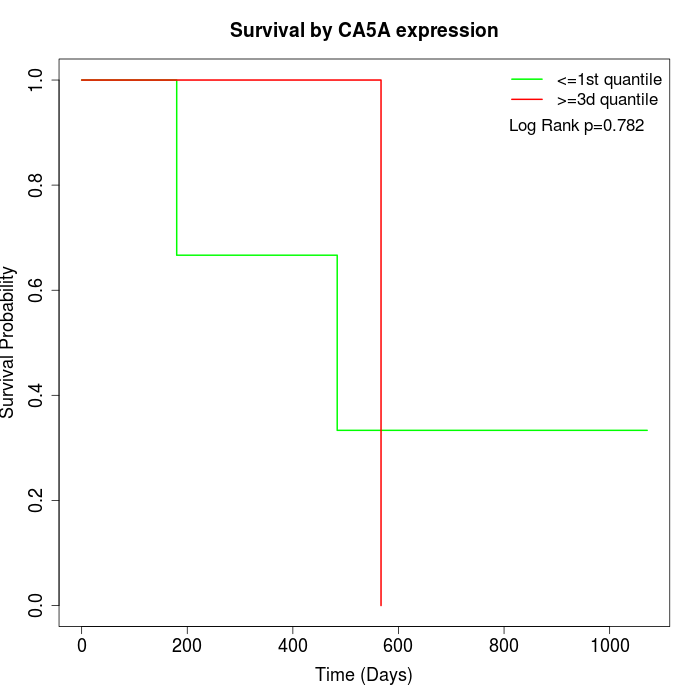
Survival by CA5A expression:
Note: Click image to view full size file.
Copy number change of CA5A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CA5A | 763 | 4 | 3 | 23 | |
| GSE20123 | CA5A | 763 | 4 | 3 | 23 | |
| GSE43470 | CA5A | 763 | 1 | 13 | 29 | |
| GSE46452 | CA5A | 763 | 38 | 1 | 20 | |
| GSE47630 | CA5A | 763 | 11 | 9 | 20 | |
| GSE54993 | CA5A | 763 | 3 | 4 | 63 | |
| GSE54994 | CA5A | 763 | 9 | 11 | 33 | |
| GSE60625 | CA5A | 763 | 4 | 0 | 7 | |
| GSE74703 | CA5A | 763 | 1 | 9 | 26 | |
| GSE74704 | CA5A | 763 | 3 | 2 | 15 | |
| TCGA | CA5A | 763 | 28 | 15 | 53 |
Total number of gains: 106; Total number of losses: 70; Total Number of normals: 312.
Somatic mutations of CA5A:
Generating mutation plots.
Highly correlated genes for CA5A:
Showing top 20/1003 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CA5A | NPHS2 | 0.745134 | 5 | 0 | 5 |
| CA5A | RGL4 | 0.712661 | 3 | 0 | 3 |
| CA5A | DDX4 | 0.704798 | 3 | 0 | 3 |
| CA5A | PRAM1 | 0.685204 | 4 | 0 | 3 |
| CA5A | RBAKDN | 0.681652 | 3 | 0 | 3 |
| CA5A | GPHB5 | 0.681637 | 3 | 0 | 3 |
| CA5A | HRC | 0.681093 | 4 | 0 | 4 |
| CA5A | RASGEF1C | 0.678898 | 3 | 0 | 3 |
| CA5A | NR1I3 | 0.668097 | 6 | 0 | 4 |
| CA5A | MYL7 | 0.667818 | 7 | 0 | 6 |
| CA5A | BRINP2 | 0.667608 | 4 | 0 | 4 |
| CA5A | PTAFR | 0.66602 | 3 | 0 | 3 |
| CA5A | USH1C | 0.664535 | 6 | 0 | 6 |
| CA5A | SEZ6L | 0.664383 | 5 | 0 | 5 |
| CA5A | SYT13 | 0.662867 | 4 | 0 | 4 |
| CA5A | CHD5 | 0.661838 | 5 | 0 | 5 |
| CA5A | TNXB | 0.661797 | 5 | 0 | 5 |
| CA5A | NKAIN4 | 0.660706 | 3 | 0 | 3 |
| CA5A | C7orf61 | 0.660685 | 3 | 0 | 3 |
| CA5A | GRM1 | 0.659974 | 6 | 0 | 6 |
For details and further investigation, click here