| Full name: sex hormone binding globulin | Alias Symbol: ABP|TEBG|MGC126834|MGC138391 | ||
| Type: protein-coding gene | Cytoband: 17p13.1 | ||
| Entrez ID: 6462 | HGNC ID: HGNC:10839 | Ensembl Gene: ENSG00000129214 | OMIM ID: 182205 |
| Drug and gene relationship at DGIdb | |||
Expression of SHBG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SHBG | 6462 | 215689_s_at | -0.0910 | 0.7896 | |
| GSE20347 | SHBG | 6462 | 215689_s_at | -0.0371 | 0.7470 | |
| GSE23400 | SHBG | 6462 | 215689_s_at | -0.2489 | 0.0000 | |
| GSE26886 | SHBG | 6462 | 215689_s_at | -0.0145 | 0.9224 | |
| GSE29001 | SHBG | 6462 | 215689_s_at | -0.2868 | 0.0886 | |
| GSE38129 | SHBG | 6462 | 215689_s_at | -0.1130 | 0.1726 | |
| GSE45670 | SHBG | 6462 | 215689_s_at | -0.0327 | 0.7931 | |
| GSE53622 | SHBG | 6462 | 52598 | -0.8905 | 0.0000 | |
| GSE53624 | SHBG | 6462 | 52598 | -0.3191 | 0.0000 | |
| GSE63941 | SHBG | 6462 | 215689_s_at | 0.2289 | 0.0941 | |
| GSE77861 | SHBG | 6462 | 215689_s_at | -0.0730 | 0.6622 | |
| TCGA | SHBG | 6462 | RNAseq | -0.0204 | 0.9754 |
Upregulated datasets: 0; Downregulated datasets: 0.
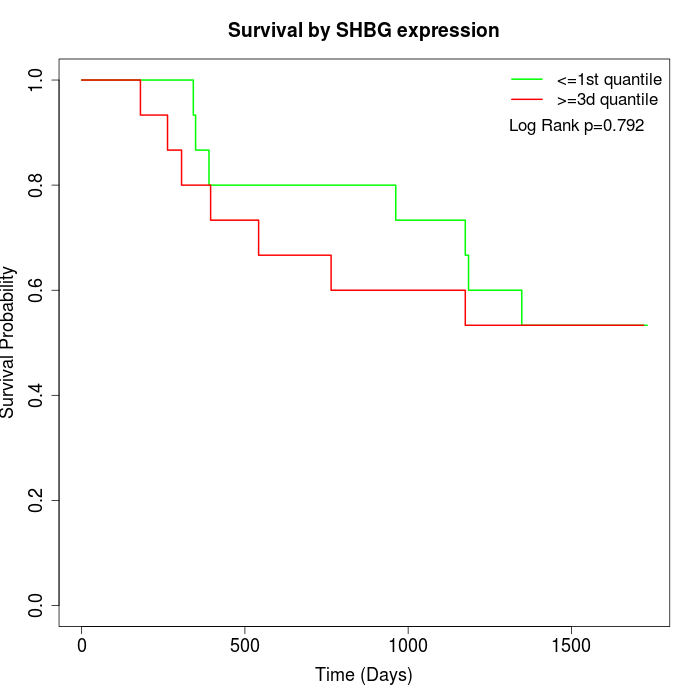
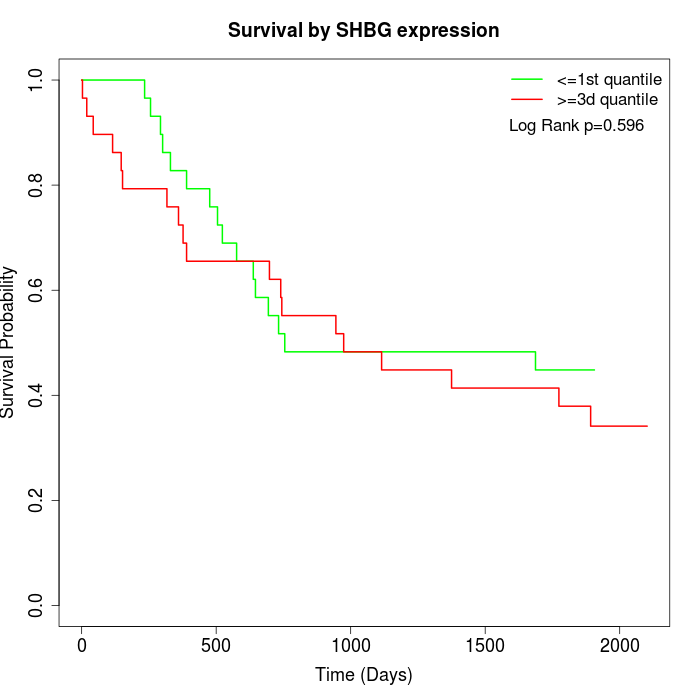
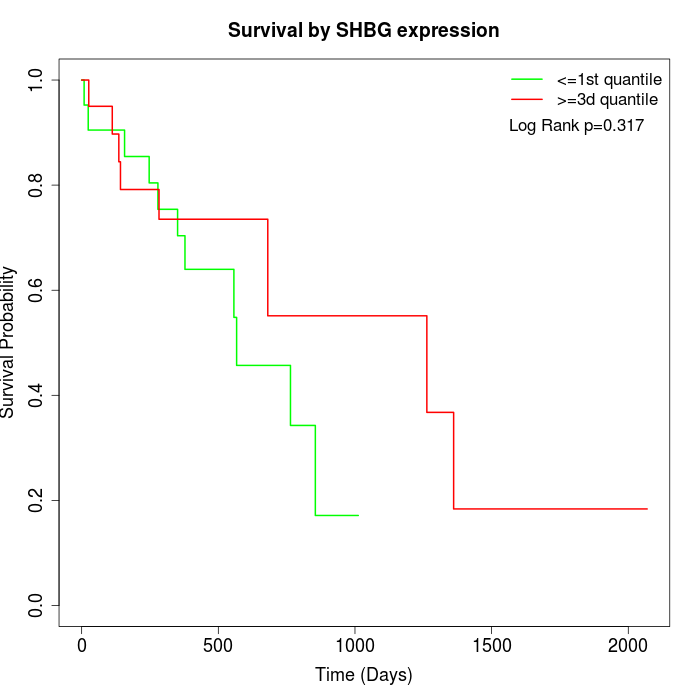
Survival by SHBG expression:
Note: Click image to view full size file.
Copy number change of SHBG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SHBG | 6462 | 4 | 2 | 24 | |
| GSE20123 | SHBG | 6462 | 4 | 3 | 23 | |
| GSE43470 | SHBG | 6462 | 1 | 6 | 36 | |
| GSE46452 | SHBG | 6462 | 34 | 1 | 24 | |
| GSE47630 | SHBG | 6462 | 7 | 1 | 32 | |
| GSE54993 | SHBG | 6462 | 4 | 3 | 63 | |
| GSE54994 | SHBG | 6462 | 5 | 8 | 40 | |
| GSE60625 | SHBG | 6462 | 4 | 0 | 7 | |
| GSE74703 | SHBG | 6462 | 1 | 3 | 32 | |
| GSE74704 | SHBG | 6462 | 2 | 1 | 17 | |
| TCGA | SHBG | 6462 | 17 | 21 | 58 |
Total number of gains: 83; Total number of losses: 49; Total Number of normals: 356.
Somatic mutations of SHBG:
Generating mutation plots.
Highly correlated genes for SHBG:
Showing top 20/1248 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SHBG | DRD5 | 0.742596 | 4 | 0 | 4 |
| SHBG | HHLA1 | 0.724272 | 8 | 0 | 8 |
| SHBG | KLF15 | 0.717097 | 4 | 0 | 4 |
| SHBG | RCVRN | 0.705133 | 5 | 0 | 5 |
| SHBG | SLC28A2 | 0.70261 | 4 | 0 | 4 |
| SHBG | NPHS2 | 0.69997 | 6 | 0 | 5 |
| SHBG | HAND1 | 0.693591 | 5 | 0 | 5 |
| SHBG | BTN1A1 | 0.689194 | 5 | 0 | 5 |
| SHBG | DLG4 | 0.688417 | 3 | 0 | 3 |
| SHBG | OVGP1 | 0.687638 | 5 | 0 | 5 |
| SHBG | CD5 | 0.685311 | 5 | 0 | 5 |
| SHBG | GJC2 | 0.679956 | 5 | 0 | 4 |
| SHBG | MTAP | 0.678944 | 5 | 0 | 5 |
| SHBG | GABRA5 | 0.67282 | 4 | 0 | 4 |
| SHBG | CAMP | 0.672625 | 4 | 0 | 4 |
| SHBG | IL1RL1 | 0.671485 | 4 | 0 | 3 |
| SHBG | NANOS2 | 0.669811 | 5 | 0 | 4 |
| SHBG | RNF39 | 0.668508 | 3 | 0 | 3 |
| SHBG | FSCN3 | 0.66685 | 5 | 0 | 4 |
| SHBG | ADAM11 | 0.666121 | 9 | 0 | 7 |
For details and further investigation, click here