| Full name: nuclear receptor subfamily 1 group I member 2 | Alias Symbol: ONR1|PXR|BXR|SXR|PAR2 | ||
| Type: protein-coding gene | Cytoband: 3q13.33 | ||
| Entrez ID: 8856 | HGNC ID: HGNC:7968 | Ensembl Gene: ENSG00000144852 | OMIM ID: 603065 |
| Drug and gene relationship at DGIdb | |||
Expression of NR1I2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NR1I2 | 8856 | 207203_s_at | -0.1202 | 0.5384 | |
| GSE20347 | NR1I2 | 8856 | 207203_s_at | 0.0510 | 0.4918 | |
| GSE23400 | NR1I2 | 8856 | 207203_s_at | -0.0377 | 0.3252 | |
| GSE26886 | NR1I2 | 8856 | 207203_s_at | 0.0456 | 0.7761 | |
| GSE29001 | NR1I2 | 8856 | 207203_s_at | -0.1227 | 0.4110 | |
| GSE38129 | NR1I2 | 8856 | 207203_s_at | -0.0360 | 0.6693 | |
| GSE45670 | NR1I2 | 8856 | 207203_s_at | -0.0330 | 0.7141 | |
| GSE53622 | NR1I2 | 8856 | 32984 | -0.2667 | 0.1576 | |
| GSE53624 | NR1I2 | 8856 | 32984 | -0.1928 | 0.1545 | |
| GSE63941 | NR1I2 | 8856 | 207203_s_at | 0.3586 | 0.0328 | |
| GSE77861 | NR1I2 | 8856 | 207203_s_at | -0.0329 | 0.7427 | |
| TCGA | NR1I2 | 8856 | RNAseq | -2.4117 | 0.0011 |
Upregulated datasets: 0; Downregulated datasets: 1.
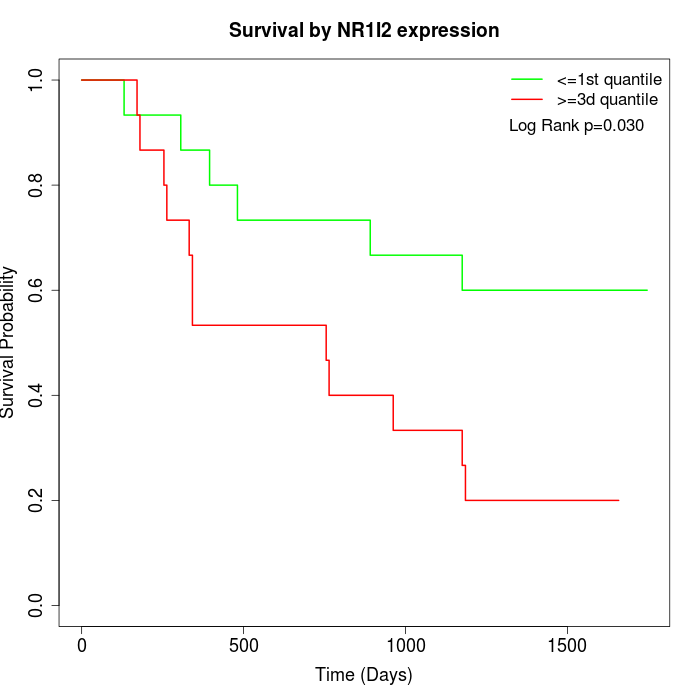
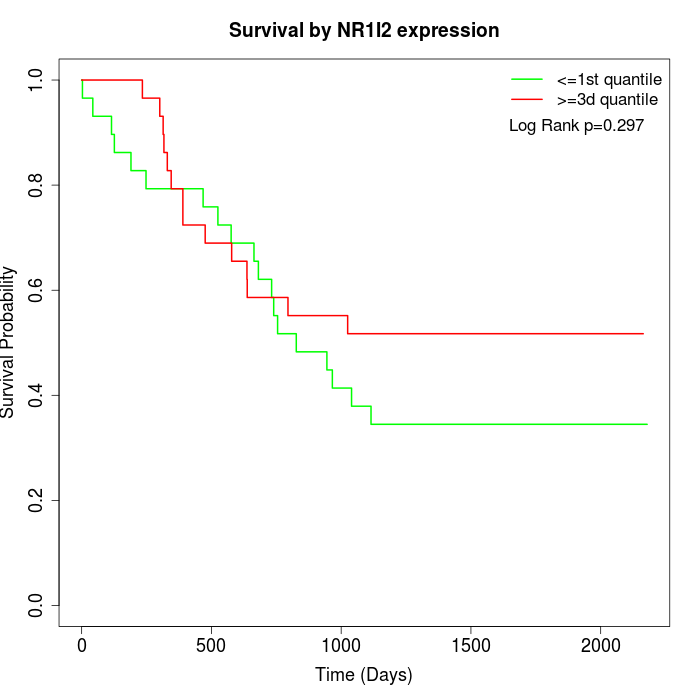
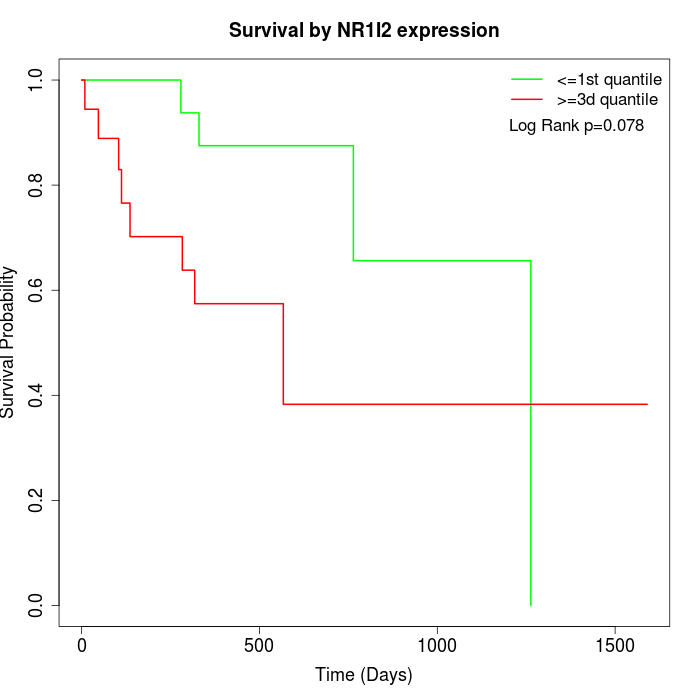
Survival by NR1I2 expression:
Note: Click image to view full size file.
Copy number change of NR1I2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NR1I2 | 8856 | 18 | 0 | 12 | |
| GSE20123 | NR1I2 | 8856 | 18 | 0 | 12 | |
| GSE43470 | NR1I2 | 8856 | 21 | 1 | 21 | |
| GSE46452 | NR1I2 | 8856 | 14 | 5 | 40 | |
| GSE47630 | NR1I2 | 8856 | 16 | 5 | 19 | |
| GSE54993 | NR1I2 | 8856 | 2 | 6 | 62 | |
| GSE54994 | NR1I2 | 8856 | 30 | 4 | 19 | |
| GSE60625 | NR1I2 | 8856 | 0 | 6 | 5 | |
| GSE74703 | NR1I2 | 8856 | 17 | 1 | 18 | |
| GSE74704 | NR1I2 | 8856 | 13 | 0 | 7 | |
| TCGA | NR1I2 | 8856 | 53 | 7 | 36 |
Total number of gains: 202; Total number of losses: 35; Total Number of normals: 251.
Somatic mutations of NR1I2:
Generating mutation plots.
Highly correlated genes for NR1I2:
Showing top 20/667 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NR1I2 | NOX1 | 0.694348 | 3 | 0 | 3 |
| NR1I2 | SAMD10 | 0.689836 | 3 | 0 | 3 |
| NR1I2 | ARMCX4 | 0.688752 | 3 | 0 | 3 |
| NR1I2 | JAK3 | 0.675463 | 4 | 0 | 4 |
| NR1I2 | HTR1D | 0.673834 | 4 | 0 | 4 |
| NR1I2 | ACR | 0.671618 | 5 | 0 | 5 |
| NR1I2 | NEUROD4 | 0.670757 | 5 | 0 | 5 |
| NR1I2 | KLK2 | 0.658212 | 4 | 0 | 4 |
| NR1I2 | SLC4A1 | 0.655021 | 6 | 0 | 6 |
| NR1I2 | LHX3 | 0.654842 | 5 | 0 | 4 |
| NR1I2 | KIAA1211L | 0.654224 | 4 | 0 | 3 |
| NR1I2 | PRELP | 0.653697 | 3 | 0 | 3 |
| NR1I2 | NTNG1 | 0.651998 | 4 | 0 | 4 |
| NR1I2 | ARSE | 0.651613 | 3 | 0 | 3 |
| NR1I2 | TECTA | 0.646797 | 4 | 0 | 3 |
| NR1I2 | TNR | 0.643207 | 5 | 0 | 4 |
| NR1I2 | KHDRBS2 | 0.640074 | 5 | 0 | 5 |
| NR1I2 | CHST5 | 0.640025 | 4 | 0 | 4 |
| NR1I2 | GFRA4 | 0.63934 | 5 | 0 | 5 |
| NR1I2 | EPOR | 0.636804 | 6 | 0 | 6 |
For details and further investigation, click here