| Full name: acrosin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 22q13.33 | ||
| Entrez ID: 49 | HGNC ID: HGNC:126 | Ensembl Gene: ENSG00000100312 | OMIM ID: 102480 |
| Drug and gene relationship at DGIdb | |||
Expression of ACR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACR | 49 | 207427_at | 0.0392 | 0.8911 | |
| GSE20347 | ACR | 49 | 207427_at | -0.0242 | 0.8246 | |
| GSE23400 | ACR | 49 | 207427_at | -0.1356 | 0.0004 | |
| GSE26886 | ACR | 49 | 207427_at | 0.0003 | 0.9983 | |
| GSE29001 | ACR | 49 | 207427_at | -0.3482 | 0.0095 | |
| GSE38129 | ACR | 49 | 207427_at | -0.1096 | 0.1815 | |
| GSE45670 | ACR | 49 | 207427_at | -0.0071 | 0.9375 | |
| GSE53622 | ACR | 49 | 139302 | 0.0153 | 0.8511 | |
| GSE53624 | ACR | 49 | 139302 | -0.1235 | 0.1012 | |
| GSE63941 | ACR | 49 | 207427_at | 0.2454 | 0.0508 | |
| GSE77861 | ACR | 49 | 207427_at | 0.0759 | 0.3488 | |
| GSE97050 | ACR | 49 | A_32_P191262 | -0.1009 | 0.6965 | |
| SRP219564 | ACR | 49 | RNAseq | -0.1690 | 0.7333 | |
| TCGA | ACR | 49 | RNAseq | 0.6127 | 0.1893 |
Upregulated datasets: 0; Downregulated datasets: 0.
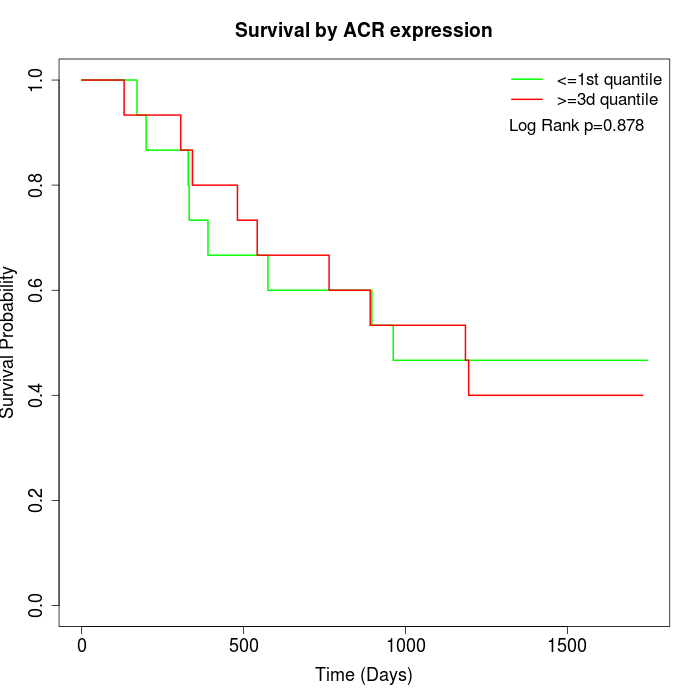
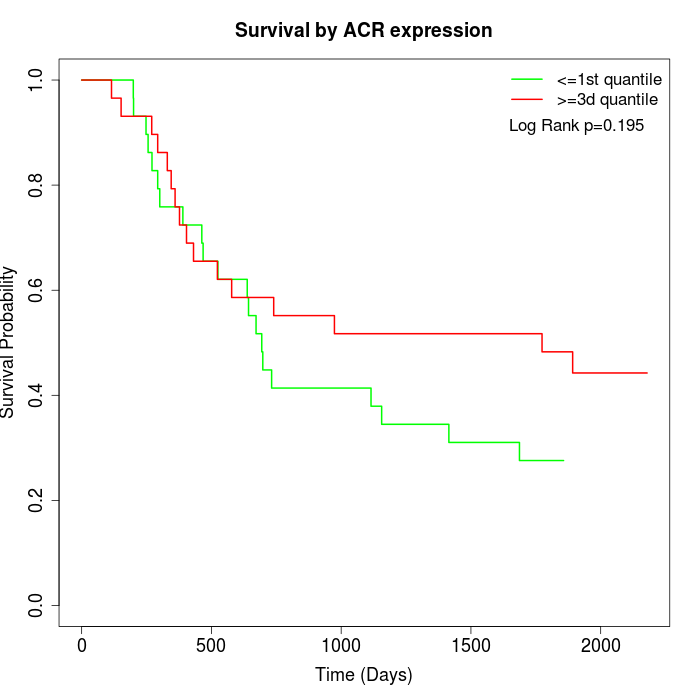
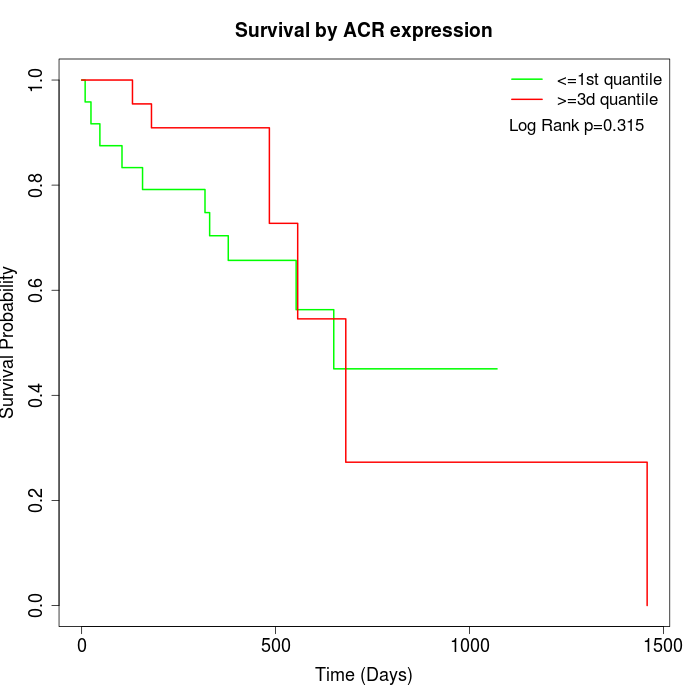
Survival by ACR expression:
Note: Click image to view full size file.
Copy number change of ACR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACR | 49 | 3 | 8 | 19 | |
| GSE20123 | ACR | 49 | 3 | 8 | 19 | |
| GSE43470 | ACR | 49 | 2 | 8 | 33 | |
| GSE46452 | ACR | 49 | 31 | 2 | 26 | |
| GSE47630 | ACR | 49 | 9 | 4 | 27 | |
| GSE54993 | ACR | 49 | 4 | 6 | 60 | |
| GSE54994 | ACR | 49 | 11 | 8 | 34 | |
| GSE60625 | ACR | 49 | 5 | 0 | 6 | |
| GSE74703 | ACR | 49 | 2 | 6 | 28 | |
| GSE74704 | ACR | 49 | 1 | 4 | 15 | |
| TCGA | ACR | 49 | 26 | 16 | 54 |
Total number of gains: 97; Total number of losses: 70; Total Number of normals: 321.
Somatic mutations of ACR:
Generating mutation plots.
Highly correlated genes for ACR:
Showing top 20/922 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACR | KATNAL2 | 0.79287 | 3 | 0 | 3 |
| ACR | OR5A1 | 0.779899 | 3 | 0 | 3 |
| ACR | DCDC1 | 0.77647 | 3 | 0 | 3 |
| ACR | DKK4 | 0.753593 | 3 | 0 | 3 |
| ACR | YY2 | 0.752694 | 3 | 0 | 3 |
| ACR | OR2T1 | 0.750594 | 3 | 0 | 3 |
| ACR | PHF7 | 0.747919 | 5 | 0 | 5 |
| ACR | STRC | 0.743992 | 3 | 0 | 3 |
| ACR | KRTAP5-9 | 0.742735 | 4 | 0 | 4 |
| ACR | RALYL | 0.739696 | 4 | 0 | 4 |
| ACR | INSC | 0.738231 | 3 | 0 | 3 |
| ACR | ABCA4 | 0.73745 | 5 | 0 | 4 |
| ACR | BPIFB4 | 0.737225 | 3 | 0 | 3 |
| ACR | RXFP3 | 0.734508 | 4 | 0 | 4 |
| ACR | CACNG4 | 0.734074 | 8 | 0 | 8 |
| ACR | GRIN1 | 0.729185 | 5 | 0 | 5 |
| ACR | POU4F3 | 0.727923 | 3 | 0 | 3 |
| ACR | KRT77 | 0.72381 | 3 | 0 | 3 |
| ACR | BRINP2 | 0.719866 | 4 | 0 | 4 |
| ACR | FSTL4 | 0.718136 | 5 | 0 | 5 |
For details and further investigation, click here