| Full name: LIM homeobox 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 8022 | HGNC ID: HGNC:6595 | Ensembl Gene: ENSG00000107187 | OMIM ID: 600577 |
| Drug and gene relationship at DGIdb | |||
Expression of LHX3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LHX3 | 8022 | 221670_s_at | 0.0676 | 0.8648 | |
| GSE20347 | LHX3 | 8022 | 221670_s_at | 0.0631 | 0.4363 | |
| GSE23400 | LHX3 | 8022 | 221670_s_at | -0.1120 | 0.0178 | |
| GSE26886 | LHX3 | 8022 | 221670_s_at | 0.1251 | 0.5126 | |
| GSE29001 | LHX3 | 8022 | 221670_s_at | -0.1171 | 0.5856 | |
| GSE38129 | LHX3 | 8022 | 221670_s_at | -0.0877 | 0.3150 | |
| GSE45670 | LHX3 | 8022 | 221670_s_at | 0.2735 | 0.0173 | |
| GSE53622 | LHX3 | 8022 | 4364 | -0.5054 | 0.0000 | |
| GSE53624 | LHX3 | 8022 | 4364 | -1.1337 | 0.0000 | |
| GSE63941 | LHX3 | 8022 | 221670_s_at | 0.3757 | 0.1164 | |
| GSE77861 | LHX3 | 8022 | 221670_s_at | 0.0434 | 0.7423 | |
| GSE97050 | LHX3 | 8022 | A_33_P3223678 | -0.0067 | 0.9812 | |
| TCGA | LHX3 | 8022 | RNAseq | 3.4149 | 0.0053 |
Upregulated datasets: 1; Downregulated datasets: 1.
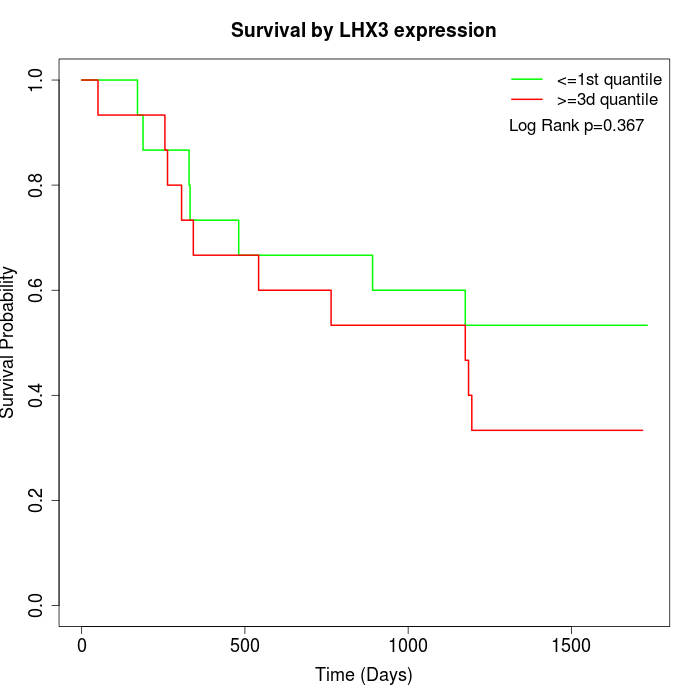
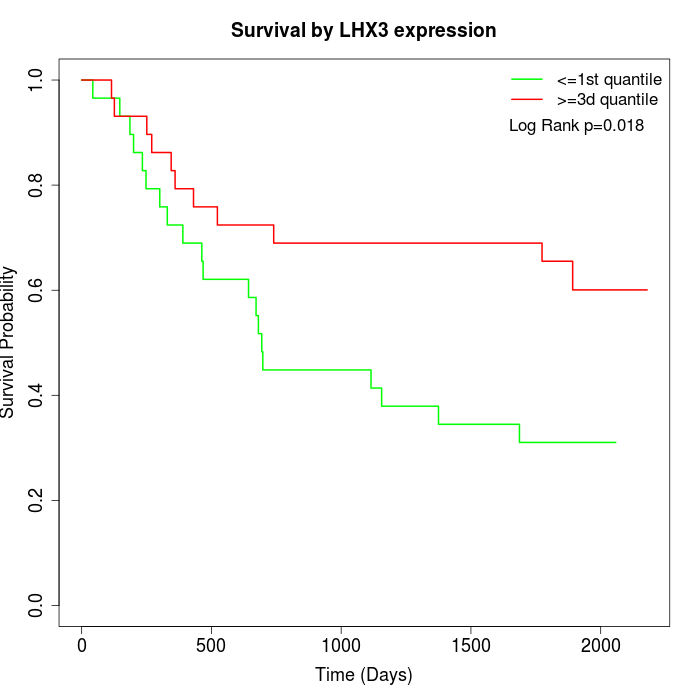
Survival by LHX3 expression:
Note: Click image to view full size file.
Copy number change of LHX3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LHX3 | 8022 | 4 | 7 | 19 | |
| GSE20123 | LHX3 | 8022 | 4 | 7 | 19 | |
| GSE43470 | LHX3 | 8022 | 3 | 7 | 33 | |
| GSE46452 | LHX3 | 8022 | 6 | 13 | 40 | |
| GSE47630 | LHX3 | 8022 | 6 | 15 | 19 | |
| GSE54993 | LHX3 | 8022 | 3 | 3 | 64 | |
| GSE54994 | LHX3 | 8022 | 12 | 8 | 33 | |
| GSE60625 | LHX3 | 8022 | 0 | 0 | 11 | |
| GSE74703 | LHX3 | 8022 | 3 | 5 | 28 | |
| GSE74704 | LHX3 | 8022 | 2 | 5 | 13 | |
| TCGA | LHX3 | 8022 | 29 | 24 | 43 |
Total number of gains: 72; Total number of losses: 94; Total Number of normals: 322.
Somatic mutations of LHX3:
Generating mutation plots.
Highly correlated genes for LHX3:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LHX3 | KIR3DL2 | 0.756692 | 3 | 0 | 3 |
| LHX3 | NELL1 | 0.732971 | 3 | 0 | 3 |
| LHX3 | KCND3 | 0.728683 | 3 | 0 | 3 |
| LHX3 | TRAF3IP2-AS1 | 0.719327 | 3 | 0 | 3 |
| LHX3 | KRTAP9-6 | 0.701113 | 3 | 0 | 3 |
| LHX3 | SFTPA2 | 0.693271 | 3 | 0 | 3 |
| LHX3 | TRIM39 | 0.69007 | 3 | 0 | 3 |
| LHX3 | CT55 | 0.68958 | 4 | 0 | 4 |
| LHX3 | KRTAP5-8 | 0.688547 | 4 | 0 | 3 |
| LHX3 | MOGAT2 | 0.683619 | 4 | 0 | 4 |
| LHX3 | VSIG8 | 0.681591 | 3 | 0 | 3 |
| LHX3 | LACTBL1 | 0.675942 | 3 | 0 | 3 |
| LHX3 | CCDC149 | 0.675373 | 5 | 0 | 5 |
| LHX3 | FABP1 | 0.675318 | 3 | 0 | 3 |
| LHX3 | SENP1 | 0.669585 | 3 | 0 | 3 |
| LHX3 | DBP | 0.666246 | 4 | 0 | 3 |
| LHX3 | ADAD2 | 0.66525 | 4 | 0 | 3 |
| LHX3 | TEAD3 | 0.664082 | 5 | 0 | 5 |
| LHX3 | SETD5 | 0.663987 | 3 | 0 | 3 |
| LHX3 | TCL6 | 0.660705 | 4 | 0 | 4 |
For details and further investigation, click here