| Full name: nuclear receptor subfamily 4 group A member 2 | Alias Symbol: TINUR|NOT|RNR1|HZF-3 | ||
| Type: protein-coding gene | Cytoband: 2q22-q23 | ||
| Entrez ID: 4929 | HGNC ID: HGNC:7981 | Ensembl Gene: ENSG00000153234 | OMIM ID: 601828 |
| Drug and gene relationship at DGIdb | |||
NR4A2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04925 | Aldosterone synthesis and secretion |
Expression of NR4A2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NR4A2 | 4929 | 216248_s_at | -0.5815 | 0.5975 | |
| GSE20347 | NR4A2 | 4929 | 204621_s_at | -0.5759 | 0.1305 | |
| GSE23400 | NR4A2 | 4929 | 204622_x_at | -0.4292 | 0.0364 | |
| GSE26886 | NR4A2 | 4929 | 204622_x_at | 0.3291 | 0.5014 | |
| GSE29001 | NR4A2 | 4929 | 204622_x_at | -0.1770 | 0.8116 | |
| GSE38129 | NR4A2 | 4929 | 204622_x_at | -0.9352 | 0.0129 | |
| GSE45670 | NR4A2 | 4929 | 204622_x_at | -0.5110 | 0.2862 | |
| GSE53622 | NR4A2 | 4929 | 67094 | -0.6760 | 0.0002 | |
| GSE53624 | NR4A2 | 4929 | 67094 | -0.3619 | 0.0001 | |
| GSE63941 | NR4A2 | 4929 | 204622_x_at | -2.6626 | 0.0071 | |
| GSE77861 | NR4A2 | 4929 | 204621_s_at | 0.0846 | 0.8879 | |
| GSE97050 | NR4A2 | 4929 | A_33_P3299066 | -0.0474 | 0.9671 | |
| SRP007169 | NR4A2 | 4929 | RNAseq | -0.2626 | 0.6819 | |
| SRP008496 | NR4A2 | 4929 | RNAseq | -0.7115 | 0.1056 | |
| SRP064894 | NR4A2 | 4929 | RNAseq | -0.6702 | 0.1170 | |
| SRP133303 | NR4A2 | 4929 | RNAseq | -0.2476 | 0.5222 | |
| SRP159526 | NR4A2 | 4929 | RNAseq | -0.0235 | 0.9487 | |
| SRP193095 | NR4A2 | 4929 | RNAseq | 0.2289 | 0.5149 | |
| SRP219564 | NR4A2 | 4929 | RNAseq | 0.4395 | 0.4516 | |
| TCGA | NR4A2 | 4929 | RNAseq | -0.4497 | 0.0004 |
Upregulated datasets: 0; Downregulated datasets: 1.
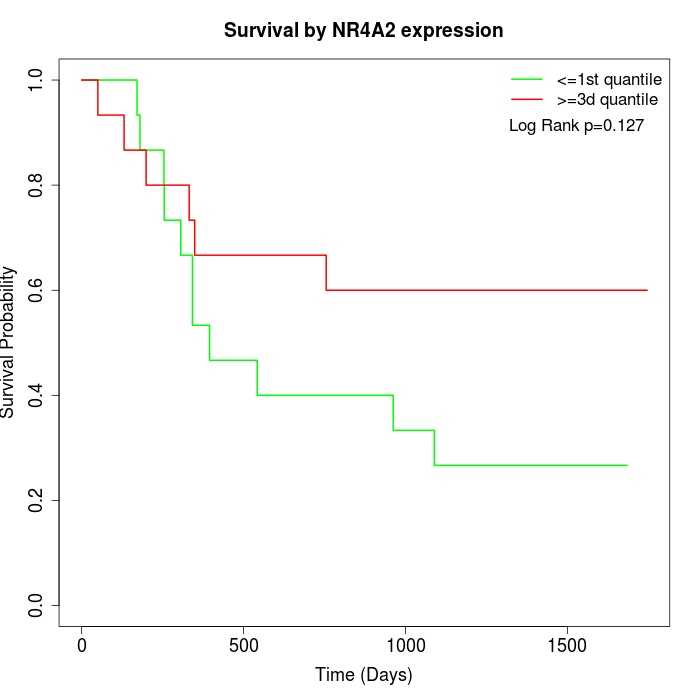
Survival by NR4A2 expression:
Note: Click image to view full size file.
Copy number change of NR4A2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NR4A2 | 4929 | 4 | 1 | 25 | |
| GSE20123 | NR4A2 | 4929 | 4 | 1 | 25 | |
| GSE43470 | NR4A2 | 4929 | 4 | 1 | 38 | |
| GSE46452 | NR4A2 | 4929 | 1 | 4 | 54 | |
| GSE47630 | NR4A2 | 4929 | 5 | 3 | 32 | |
| GSE54993 | NR4A2 | 4929 | 0 | 4 | 66 | |
| GSE54994 | NR4A2 | 4929 | 11 | 2 | 40 | |
| GSE60625 | NR4A2 | 4929 | 0 | 3 | 8 | |
| GSE74703 | NR4A2 | 4929 | 3 | 1 | 32 | |
| GSE74704 | NR4A2 | 4929 | 3 | 0 | 17 | |
| TCGA | NR4A2 | 4929 | 21 | 9 | 66 |
Total number of gains: 56; Total number of losses: 29; Total Number of normals: 403.
Somatic mutations of NR4A2:
Generating mutation plots.
Highly correlated genes for NR4A2:
Showing top 20/262 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NR4A2 | GPR3 | 0.762243 | 3 | 0 | 3 |
| NR4A2 | SOX17 | 0.690936 | 5 | 0 | 4 |
| NR4A2 | FOSB | 0.68909 | 11 | 0 | 10 |
| NR4A2 | ACTB | 0.687264 | 3 | 0 | 3 |
| NR4A2 | NR4A1 | 0.676094 | 9 | 0 | 7 |
| NR4A2 | GKN1 | 0.676066 | 3 | 0 | 3 |
| NR4A2 | IER2 | 0.674884 | 7 | 0 | 6 |
| NR4A2 | LIMS1 | 0.664978 | 3 | 0 | 3 |
| NR4A2 | C11orf96 | 0.663974 | 4 | 0 | 3 |
| NR4A2 | NFE4 | 0.661559 | 3 | 0 | 3 |
| NR4A2 | AFMID | 0.661304 | 3 | 0 | 3 |
| NR4A2 | DNAJC18 | 0.655711 | 3 | 0 | 3 |
| NR4A2 | ANKRD34C | 0.654203 | 3 | 0 | 3 |
| NR4A2 | TBX5 | 0.653806 | 3 | 0 | 3 |
| NR4A2 | DUSP6 | 0.650708 | 3 | 0 | 3 |
| NR4A2 | NR4A3 | 0.650405 | 12 | 0 | 11 |
| NR4A2 | LRCH1 | 0.646944 | 4 | 0 | 3 |
| NR4A2 | DNAJB4 | 0.645772 | 7 | 0 | 7 |
| NR4A2 | CYP4V2 | 0.644824 | 3 | 0 | 3 |
| NR4A2 | HDAC9 | 0.642049 | 3 | 0 | 3 |
For details and further investigation, click here