| Full name: gastrokine 1 | Alias Symbol: AMP18|CA11|BRICD1 | ||
| Type: protein-coding gene | Cytoband: 2p13.3 | ||
| Entrez ID: 56287 | HGNC ID: HGNC:23217 | Ensembl Gene: ENSG00000169605 | OMIM ID: 606402 |
| Drug and gene relationship at DGIdb | |||
Expression of GKN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GKN1 | 56287 | 220191_at | -0.1477 | 0.7017 | |
| GSE20347 | GKN1 | 56287 | 220191_at | 0.0688 | 0.2162 | |
| GSE23400 | GKN1 | 56287 | 220191_at | -0.3532 | 0.1411 | |
| GSE26886 | GKN1 | 56287 | 220191_at | -0.3972 | 0.2858 | |
| GSE29001 | GKN1 | 56287 | 220191_at | -0.0949 | 0.4637 | |
| GSE38129 | GKN1 | 56287 | 220191_at | -0.9323 | 0.1102 | |
| GSE45670 | GKN1 | 56287 | 220191_at | 0.0424 | 0.6028 | |
| GSE53622 | GKN1 | 56287 | 36072 | 0.1017 | 0.8399 | |
| GSE53624 | GKN1 | 56287 | 36072 | -0.7508 | 0.0595 | |
| GSE63941 | GKN1 | 56287 | 220191_at | 0.0290 | 0.8503 | |
| GSE77861 | GKN1 | 56287 | 220191_at | 0.0163 | 0.8753 | |
| TCGA | GKN1 | 56287 | RNAseq | -6.3147 | 0.0004 |
Upregulated datasets: 0; Downregulated datasets: 1.
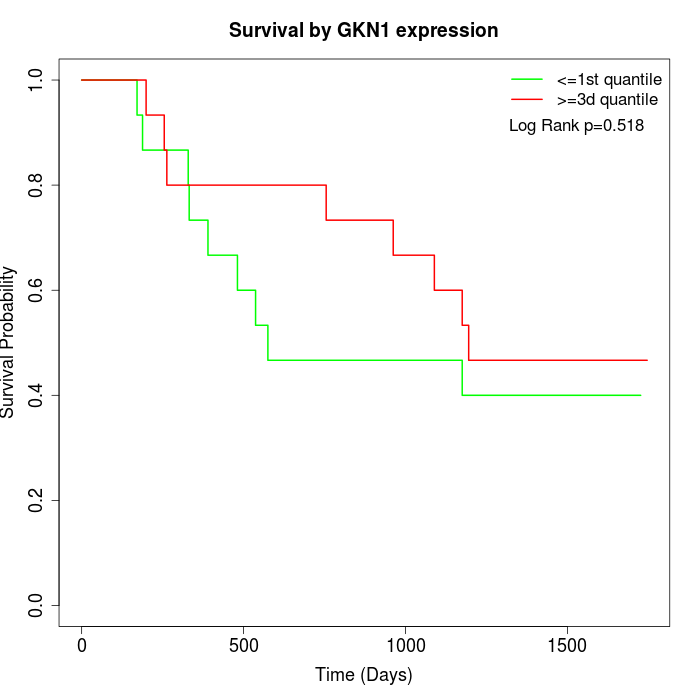
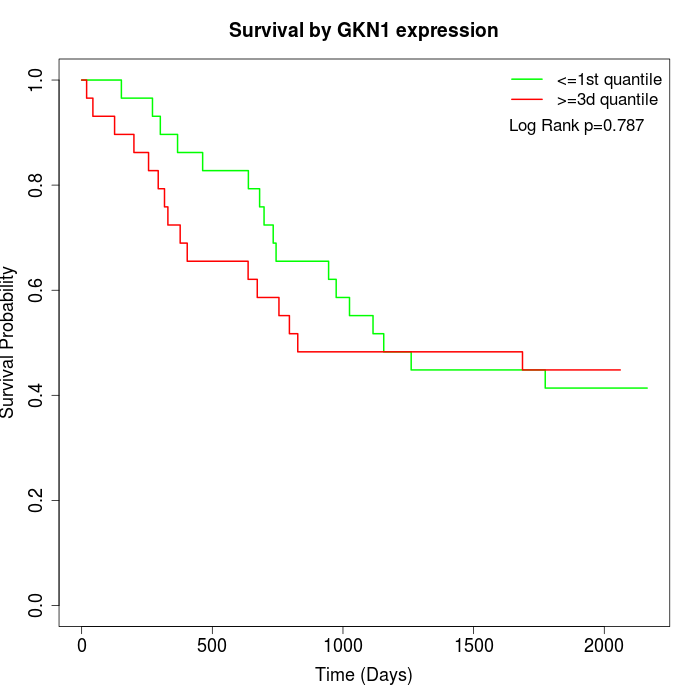
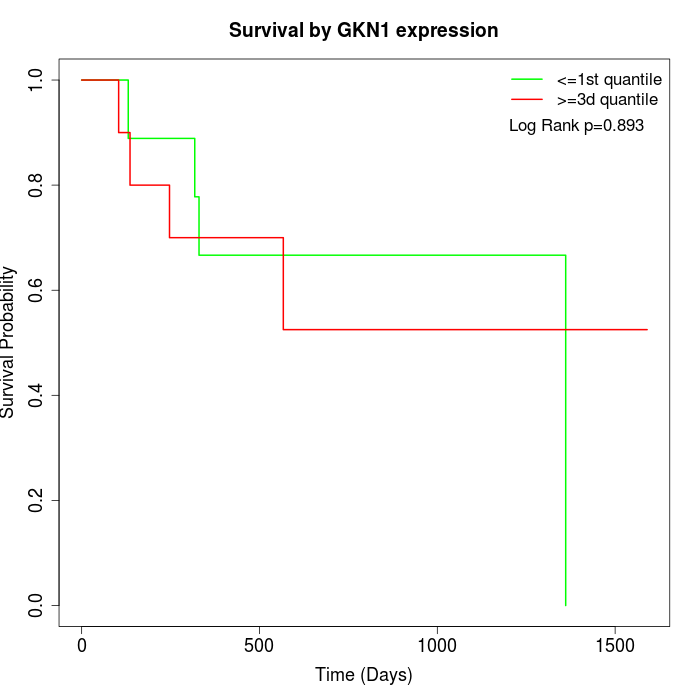
Survival by GKN1 expression:
Note: Click image to view full size file.
Copy number change of GKN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GKN1 | 56287 | 11 | 0 | 19 | |
| GSE20123 | GKN1 | 56287 | 11 | 0 | 19 | |
| GSE43470 | GKN1 | 56287 | 6 | 0 | 37 | |
| GSE46452 | GKN1 | 56287 | 2 | 3 | 54 | |
| GSE47630 | GKN1 | 56287 | 7 | 0 | 33 | |
| GSE54993 | GKN1 | 56287 | 0 | 6 | 64 | |
| GSE54994 | GKN1 | 56287 | 12 | 0 | 41 | |
| GSE60625 | GKN1 | 56287 | 0 | 3 | 8 | |
| GSE74703 | GKN1 | 56287 | 6 | 0 | 30 | |
| GSE74704 | GKN1 | 56287 | 9 | 0 | 11 | |
| TCGA | GKN1 | 56287 | 36 | 2 | 58 |
Total number of gains: 100; Total number of losses: 14; Total Number of normals: 374.
Somatic mutations of GKN1:
Generating mutation plots.
Highly correlated genes for GKN1:
Showing top 20/103 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GKN1 | LIPF | 0.790659 | 7 | 0 | 7 |
| GKN1 | KRT20 | 0.786674 | 4 | 0 | 4 |
| GKN1 | PGC | 0.779316 | 6 | 0 | 6 |
| GKN1 | SPINK1 | 0.772076 | 5 | 0 | 5 |
| GKN1 | CLDN18 | 0.764026 | 7 | 0 | 7 |
| GKN1 | GKN2 | 0.706692 | 5 | 0 | 4 |
| GKN1 | TFF1 | 0.699392 | 7 | 0 | 6 |
| GKN1 | RNF122 | 0.692884 | 3 | 0 | 3 |
| GKN1 | FUT9 | 0.67796 | 5 | 0 | 5 |
| GKN1 | NR4A2 | 0.676066 | 3 | 0 | 3 |
| GKN1 | REG1A | 0.675195 | 5 | 0 | 5 |
| GKN1 | MUC6 | 0.668875 | 7 | 0 | 7 |
| GKN1 | FBP2 | 0.660294 | 4 | 0 | 3 |
| GKN1 | GC | 0.653966 | 5 | 0 | 4 |
| GKN1 | CTSE | 0.651126 | 7 | 0 | 4 |
| GKN1 | CHIA | 0.65107 | 7 | 0 | 5 |
| GKN1 | NR0B2 | 0.647758 | 4 | 0 | 3 |
| GKN1 | LGALS4 | 0.639781 | 7 | 0 | 6 |
| GKN1 | ATP4A | 0.638254 | 7 | 0 | 6 |
| GKN1 | TFF2 | 0.638148 | 6 | 0 | 4 |
For details and further investigation, click here