| Full name: phosphodiesterase 2A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q13.4 | ||
| Entrez ID: 5138 | HGNC ID: HGNC:8777 | Ensembl Gene: ENSG00000186642 | OMIM ID: 602658 |
| Drug and gene relationship at DGIdb | |||
PDE2A involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04740 | Olfactory transduction | |
| hsa04925 | Aldosterone synthesis and secretion |
Expression of PDE2A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE2A | 5138 | 204134_at | -1.0856 | 0.0226 | |
| GSE20347 | PDE2A | 5138 | 204134_at | -0.4581 | 0.0001 | |
| GSE23400 | PDE2A | 5138 | 204134_at | -0.7501 | 0.0000 | |
| GSE26886 | PDE2A | 5138 | 204134_at | -0.2892 | 0.2053 | |
| GSE29001 | PDE2A | 5138 | 204134_at | -0.6893 | 0.0158 | |
| GSE38129 | PDE2A | 5138 | 204134_at | -1.3042 | 0.0000 | |
| GSE45670 | PDE2A | 5138 | 204134_at | -1.3565 | 0.0000 | |
| GSE53622 | PDE2A | 5138 | 81702 | -1.8979 | 0.0000 | |
| GSE53624 | PDE2A | 5138 | 93731 | -1.2782 | 0.0000 | |
| GSE63941 | PDE2A | 5138 | 204134_at | 0.3057 | 0.1530 | |
| GSE77861 | PDE2A | 5138 | 204134_at | -0.0240 | 0.8569 | |
| GSE97050 | PDE2A | 5138 | A_23_P401106 | -0.5996 | 0.1404 | |
| SRP007169 | PDE2A | 5138 | RNAseq | -0.8231 | 0.1382 | |
| SRP064894 | PDE2A | 5138 | RNAseq | -0.5550 | 0.1586 | |
| SRP133303 | PDE2A | 5138 | RNAseq | -2.0728 | 0.0000 | |
| SRP159526 | PDE2A | 5138 | RNAseq | -1.6905 | 0.0027 | |
| SRP193095 | PDE2A | 5138 | RNAseq | -0.4726 | 0.0894 | |
| SRP219564 | PDE2A | 5138 | RNAseq | -0.6641 | 0.0503 | |
| TCGA | PDE2A | 5138 | RNAseq | -1.0121 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 8.
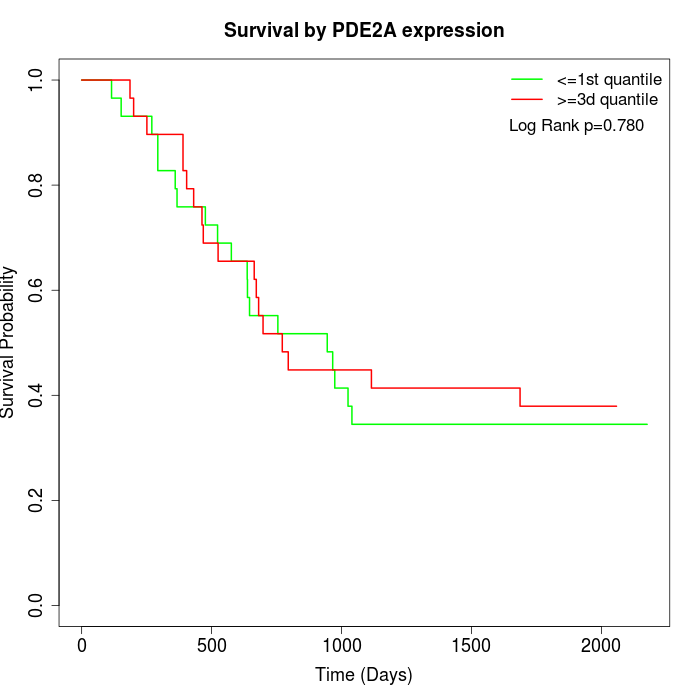
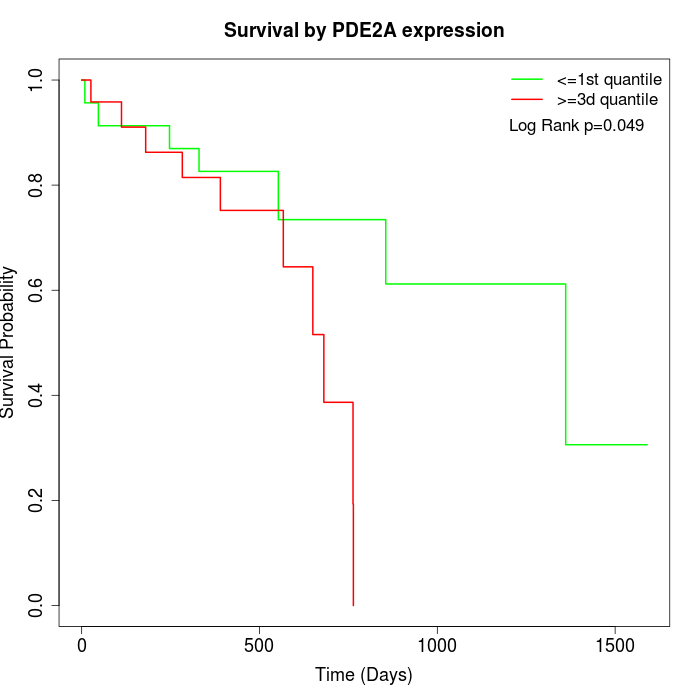
Survival by PDE2A expression:
Note: Click image to view full size file.
Copy number change of PDE2A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE2A | 5138 | 9 | 7 | 14 | |
| GSE20123 | PDE2A | 5138 | 9 | 7 | 14 | |
| GSE43470 | PDE2A | 5138 | 9 | 3 | 31 | |
| GSE46452 | PDE2A | 5138 | 11 | 18 | 30 | |
| GSE47630 | PDE2A | 5138 | 9 | 11 | 20 | |
| GSE54993 | PDE2A | 5138 | 8 | 2 | 60 | |
| GSE54994 | PDE2A | 5138 | 9 | 15 | 29 | |
| GSE60625 | PDE2A | 5138 | 0 | 7 | 4 | |
| GSE74703 | PDE2A | 5138 | 7 | 2 | 27 | |
| GSE74704 | PDE2A | 5138 | 6 | 4 | 10 | |
| TCGA | PDE2A | 5138 | 36 | 21 | 39 |
Total number of gains: 113; Total number of losses: 97; Total Number of normals: 278.
Somatic mutations of PDE2A:
Generating mutation plots.
Highly correlated genes for PDE2A:
Showing top 20/1182 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE2A | CLEC3B | 0.797461 | 3 | 0 | 3 |
| PDE2A | FXYD1 | 0.774766 | 7 | 0 | 7 |
| PDE2A | CYP21A2 | 0.77414 | 3 | 0 | 3 |
| PDE2A | CCL14 | 0.760214 | 3 | 0 | 3 |
| PDE2A | PGM5 | 0.754433 | 9 | 0 | 9 |
| PDE2A | LINC01279 | 0.753565 | 3 | 0 | 3 |
| PDE2A | CHRDL1 | 0.747405 | 8 | 0 | 8 |
| PDE2A | HSPB2 | 0.737536 | 9 | 0 | 9 |
| PDE2A | PLAC9 | 0.731395 | 5 | 0 | 5 |
| PDE2A | KLHL30 | 0.731024 | 3 | 0 | 3 |
| PDE2A | TEK | 0.722314 | 8 | 0 | 8 |
| PDE2A | ATP1A2 | 0.721263 | 9 | 0 | 8 |
| PDE2A | C1QTNF7 | 0.719601 | 5 | 0 | 5 |
| PDE2A | MYZAP | 0.718765 | 3 | 0 | 3 |
| PDE2A | ACKR1 | 0.718097 | 10 | 0 | 8 |
| PDE2A | RBFOX3 | 0.716506 | 3 | 0 | 3 |
| PDE2A | RERGL | 0.71242 | 9 | 0 | 8 |
| PDE2A | CDH19 | 0.711959 | 8 | 0 | 8 |
| PDE2A | MOB2 | 0.710135 | 3 | 0 | 3 |
| PDE2A | ACOX2 | 0.709714 | 9 | 0 | 8 |
For details and further investigation, click here