| Full name: C-C motif chemokine ligand 14 | Alias Symbol: HCC-1|HCC-3|NCC-2|SCYL2|CKb1|MCIF | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 6358 | HGNC ID: HGNC:10612 | Ensembl Gene: ENSG00000276409 | OMIM ID: 601392 |
| Drug and gene relationship at DGIdb | |||
CCL14 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CCL14 | 6358 | 24459 | -2.4568 | 0.0000 | |
| GSE53624 | CCL14 | 6358 | 24459 | -2.3039 | 0.0000 | |
| GSE97050 | CCL14 | 6358 | A_23_P218369 | -0.4818 | 0.3302 | |
| SRP064894 | CCL14 | 6358 | RNAseq | -0.1659 | 0.8213 | |
| SRP133303 | CCL14 | 6358 | RNAseq | -2.7993 | 0.0076 | |
| SRP159526 | CCL14 | 6358 | RNAseq | -2.3334 | 0.0168 | |
| SRP219564 | CCL14 | 6358 | RNAseq | -0.9687 | 0.4869 | |
| TCGA | CCL14 | 6358 | RNAseq | -1.6501 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 5.
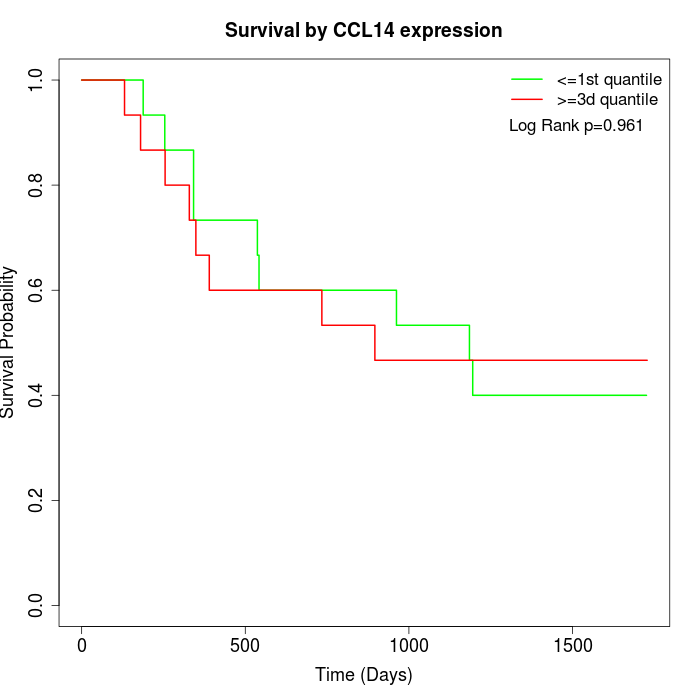
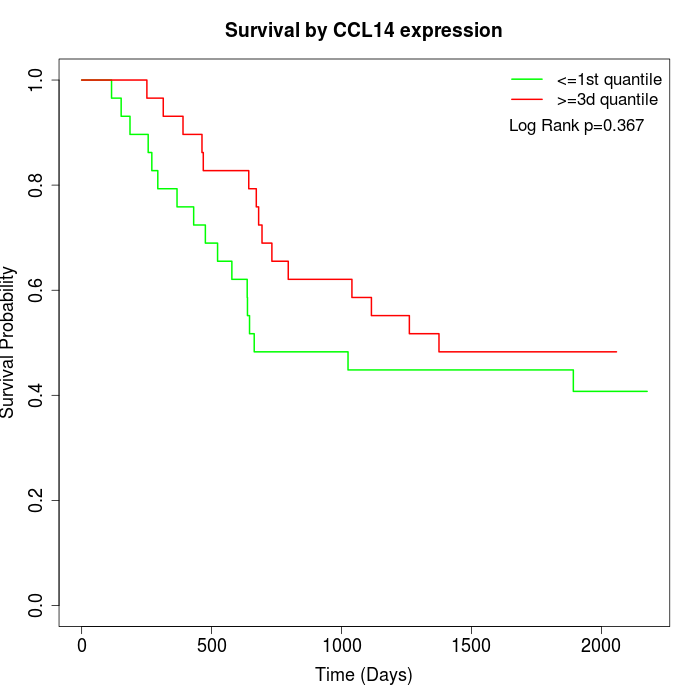
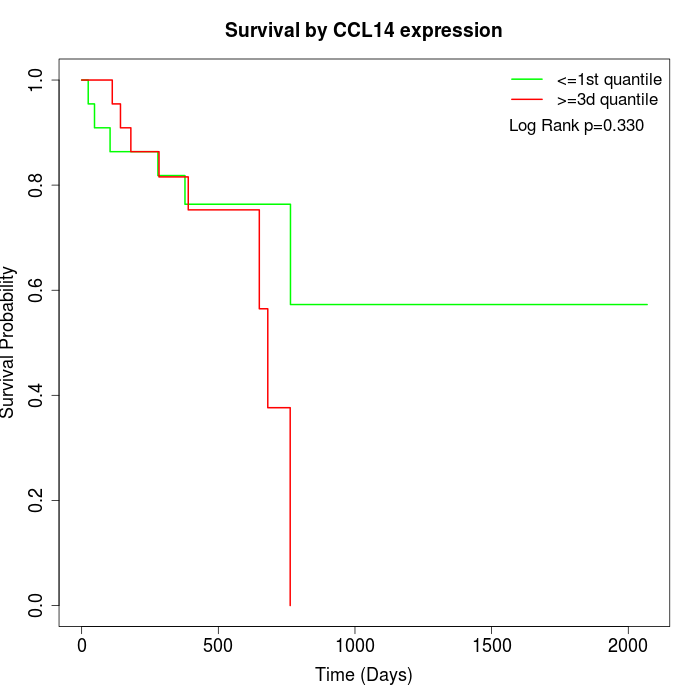
Survival by CCL14 expression:
Note: Click image to view full size file.
Copy number change of CCL14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL14 | 6358 | 5 | 1 | 24 | |
| GSE20123 | CCL14 | 6358 | 5 | 1 | 24 | |
| GSE43470 | CCL14 | 6358 | 1 | 2 | 40 | |
| GSE46452 | CCL14 | 6358 | 34 | 0 | 25 | |
| GSE47630 | CCL14 | 6358 | 7 | 1 | 32 | |
| GSE54993 | CCL14 | 6358 | 3 | 3 | 64 | |
| GSE54994 | CCL14 | 6358 | 8 | 6 | 39 | |
| GSE60625 | CCL14 | 6358 | 4 | 0 | 7 | |
| GSE74703 | CCL14 | 6358 | 1 | 1 | 34 | |
| GSE74704 | CCL14 | 6358 | 3 | 1 | 16 | |
| TCGA | CCL14 | 6358 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of CCL14:
Generating mutation plots.
Highly correlated genes for CCL14:
Showing top 20/650 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL14 | CFD | 0.842496 | 3 | 0 | 3 |
| CCL14 | MYRIP | 0.837851 | 3 | 0 | 3 |
| CCL14 | CMA1 | 0.826397 | 3 | 0 | 3 |
| CCL14 | CYP21A2 | 0.824391 | 3 | 0 | 3 |
| CCL14 | LYVE1 | 0.824127 | 3 | 0 | 3 |
| CCL14 | ASPA | 0.820919 | 3 | 0 | 3 |
| CCL14 | RCAN2 | 0.81738 | 3 | 0 | 3 |
| CCL14 | PI16 | 0.813074 | 3 | 0 | 3 |
| CCL14 | ANGPTL1 | 0.807565 | 3 | 0 | 3 |
| CCL14 | F10 | 0.800041 | 3 | 0 | 3 |
| CCL14 | COL14A1 | 0.796272 | 3 | 0 | 3 |
| CCL14 | NOSTRIN | 0.792274 | 3 | 0 | 3 |
| CCL14 | SORCS1 | 0.789711 | 3 | 0 | 3 |
| CCL14 | C1QTNF7 | 0.784916 | 3 | 0 | 3 |
| CCL14 | ABCA8 | 0.781718 | 3 | 0 | 3 |
| CCL14 | RERGL | 0.780881 | 3 | 0 | 3 |
| CCL14 | CADM3 | 0.780638 | 3 | 0 | 3 |
| CCL14 | EBF1 | 0.775066 | 3 | 0 | 3 |
| CCL14 | PPM1M | 0.767628 | 3 | 0 | 3 |
| CCL14 | ITIH5 | 0.766668 | 3 | 0 | 3 |
For details and further investigation, click here