| Full name: kelch like family member 30 | Alias Symbol: FLJ43374 | ||
| Type: protein-coding gene | Cytoband: 2q37.3 | ||
| Entrez ID: 377007 | HGNC ID: HGNC:24770 | Ensembl Gene: ENSG00000168427 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL30:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | KLHL30 | 377007 | 15613 | -1.2853 | 0.0000 | |
| GSE53624 | KLHL30 | 377007 | 15613 | -1.2291 | 0.0000 | |
| GSE97050 | KLHL30 | 377007 | A_33_P3243702 | -0.5752 | 0.2726 | |
| SRP064894 | KLHL30 | 377007 | RNAseq | -0.5709 | 0.0617 | |
| SRP133303 | KLHL30 | 377007 | RNAseq | -0.3037 | 0.4837 | |
| SRP159526 | KLHL30 | 377007 | RNAseq | -1.0194 | 0.1730 | |
| SRP219564 | KLHL30 | 377007 | RNAseq | -0.7189 | 0.4200 | |
| TCGA | KLHL30 | 377007 | RNAseq | -1.3769 | 0.0009 |
Upregulated datasets: 0; Downregulated datasets: 3.
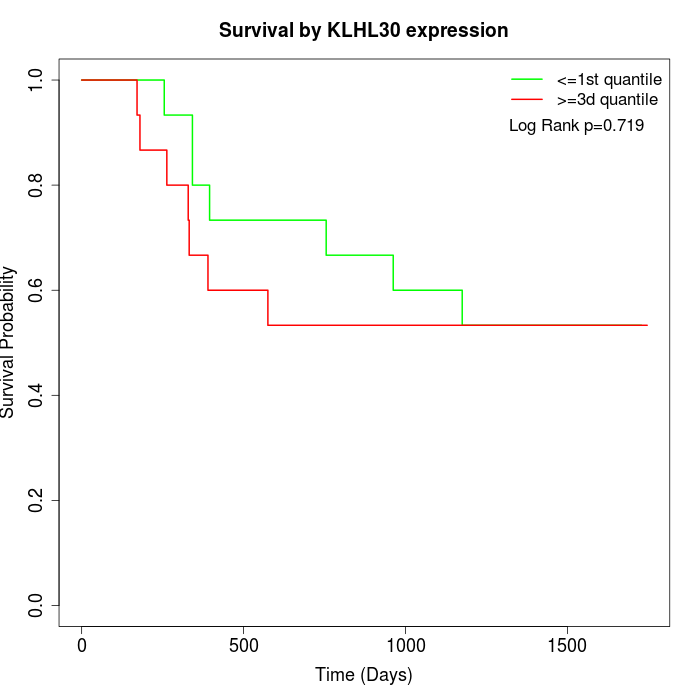
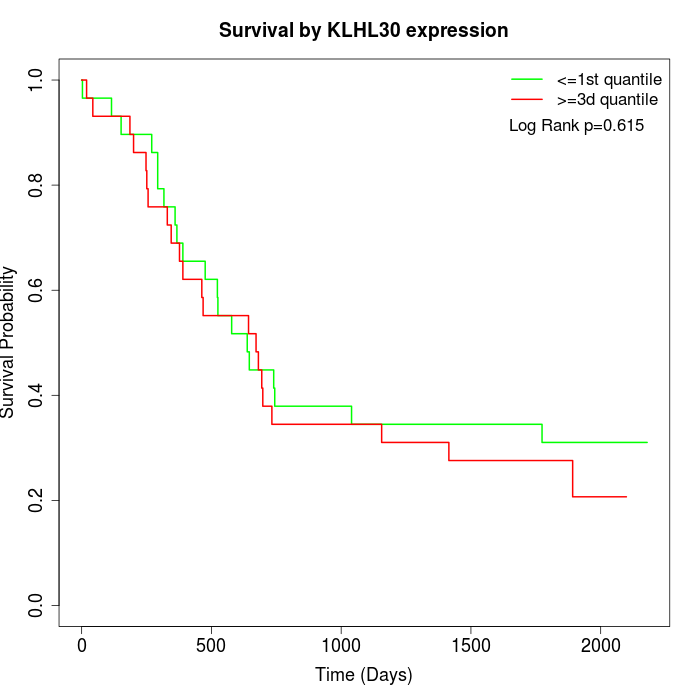
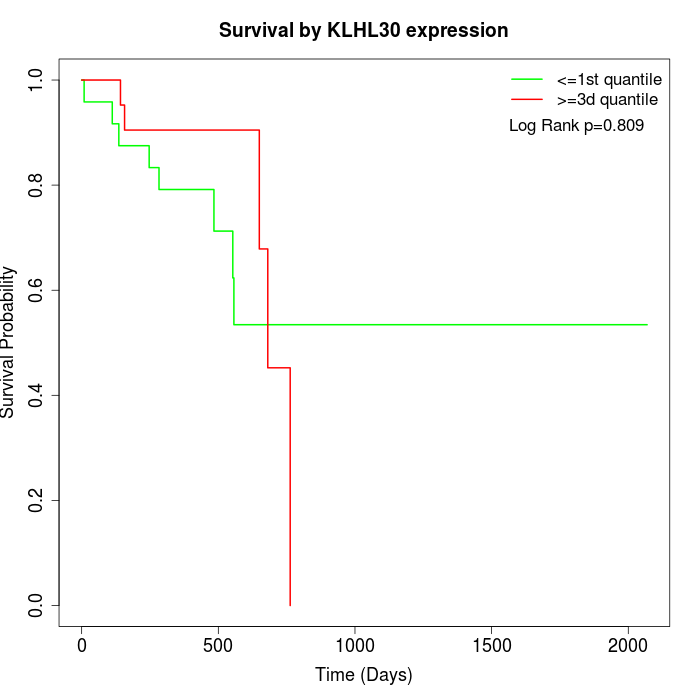
Survival by KLHL30 expression:
Note: Click image to view full size file.
Copy number change of KLHL30:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL30 | 377007 | 1 | 12 | 17 | |
| GSE20123 | KLHL30 | 377007 | 1 | 12 | 17 | |
| GSE43470 | KLHL30 | 377007 | 1 | 9 | 33 | |
| GSE46452 | KLHL30 | 377007 | 0 | 5 | 54 | |
| GSE47630 | KLHL30 | 377007 | 4 | 5 | 31 | |
| GSE54993 | KLHL30 | 377007 | 3 | 2 | 65 | |
| GSE54994 | KLHL30 | 377007 | 8 | 9 | 36 | |
| GSE60625 | KLHL30 | 377007 | 0 | 3 | 8 | |
| GSE74703 | KLHL30 | 377007 | 1 | 7 | 28 | |
| GSE74704 | KLHL30 | 377007 | 1 | 5 | 14 | |
| TCGA | KLHL30 | 377007 | 12 | 27 | 57 |
Total number of gains: 32; Total number of losses: 96; Total Number of normals: 360.
Somatic mutations of KLHL30:
Generating mutation plots.
Highly correlated genes for KLHL30:
Showing top 20/292 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL30 | PTH1R | 0.777236 | 3 | 0 | 3 |
| KLHL30 | SH3BGR | 0.76759 | 3 | 0 | 3 |
| KLHL30 | CBX7 | 0.76416 | 3 | 0 | 3 |
| KLHL30 | C3orf70 | 0.755646 | 3 | 0 | 3 |
| KLHL30 | NEGR1 | 0.753489 | 3 | 0 | 3 |
| KLHL30 | PYGM | 0.753129 | 3 | 0 | 3 |
| KLHL30 | CCDC69 | 0.752039 | 3 | 0 | 3 |
| KLHL30 | BHMT2 | 0.750217 | 3 | 0 | 3 |
| KLHL30 | F10 | 0.749697 | 3 | 0 | 3 |
| KLHL30 | SORCS1 | 0.749043 | 3 | 0 | 3 |
| KLHL30 | RCAN2 | 0.748662 | 3 | 0 | 3 |
| KLHL30 | ARHGAP6 | 0.748008 | 3 | 0 | 3 |
| KLHL30 | ANGPTL7 | 0.747361 | 3 | 0 | 3 |
| KLHL30 | ITGA7 | 0.746062 | 3 | 0 | 3 |
| KLHL30 | SGCA | 0.744847 | 3 | 0 | 3 |
| KLHL30 | CADM3 | 0.741989 | 3 | 0 | 3 |
| KLHL30 | TCEAL1 | 0.741031 | 3 | 0 | 3 |
| KLHL30 | MOAP1 | 0.740648 | 3 | 0 | 3 |
| KLHL30 | CKMT2 | 0.739975 | 3 | 0 | 3 |
| KLHL30 | CASQ2 | 0.739722 | 3 | 0 | 3 |
For details and further investigation, click here