| Full name: plexin B3 | Alias Symbol: PLEXR|PLEXB3 | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 5365 | HGNC ID: HGNC:9105 | Ensembl Gene: ENSG00000198753 | OMIM ID: 300214 |
| Drug and gene relationship at DGIdb | |||
PLXNB3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of PLXNB3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLXNB3 | 5365 | 205957_at | 0.1730 | 0.6478 | |
| GSE20347 | PLXNB3 | 5365 | 205957_at | 0.3652 | 0.0000 | |
| GSE23400 | PLXNB3 | 5365 | 205957_at | -0.0181 | 0.7990 | |
| GSE26886 | PLXNB3 | 5365 | 205957_at | 0.1158 | 0.3173 | |
| GSE29001 | PLXNB3 | 5365 | 205957_at | 0.0414 | 0.8399 | |
| GSE38129 | PLXNB3 | 5365 | 205957_at | 0.2489 | 0.0209 | |
| GSE45670 | PLXNB3 | 5365 | 205957_at | 0.2483 | 0.0224 | |
| GSE53622 | PLXNB3 | 5365 | 35053 | 0.1267 | 0.0693 | |
| GSE53624 | PLXNB3 | 5365 | 35053 | 0.1260 | 0.2726 | |
| GSE63941 | PLXNB3 | 5365 | 205957_at | 0.2980 | 0.1070 | |
| GSE77861 | PLXNB3 | 5365 | 205957_at | 0.1149 | 0.5363 | |
| GSE97050 | PLXNB3 | 5365 | A_33_P3413038 | -0.1800 | 0.7076 | |
| SRP007169 | PLXNB3 | 5365 | RNAseq | 2.5872 | 0.0022 | |
| SRP064894 | PLXNB3 | 5365 | RNAseq | 1.0184 | 0.0003 | |
| SRP133303 | PLXNB3 | 5365 | RNAseq | 1.1450 | 0.0006 | |
| SRP159526 | PLXNB3 | 5365 | RNAseq | 0.8432 | 0.0122 | |
| SRP193095 | PLXNB3 | 5365 | RNAseq | 1.6520 | 0.0000 | |
| SRP219564 | PLXNB3 | 5365 | RNAseq | 0.6178 | 0.2937 | |
| TCGA | PLXNB3 | 5365 | RNAseq | 0.2684 | 0.0065 |
Upregulated datasets: 4; Downregulated datasets: 0.
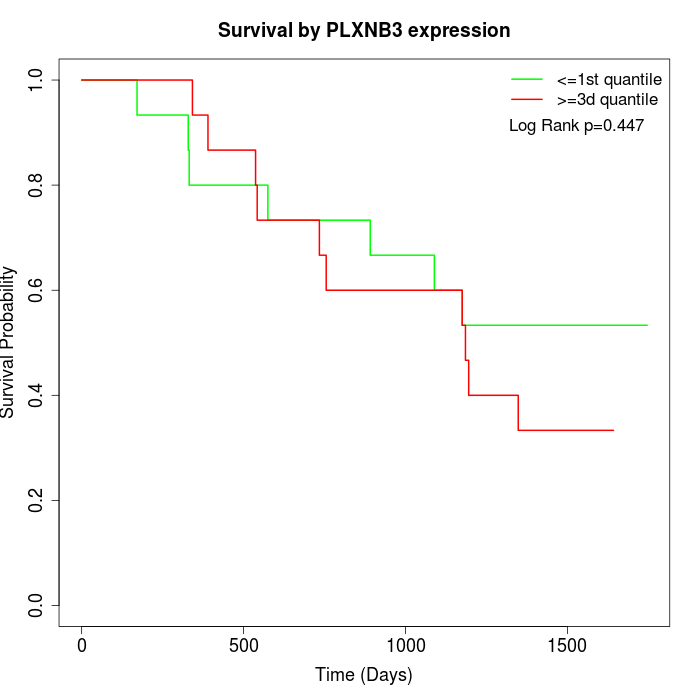
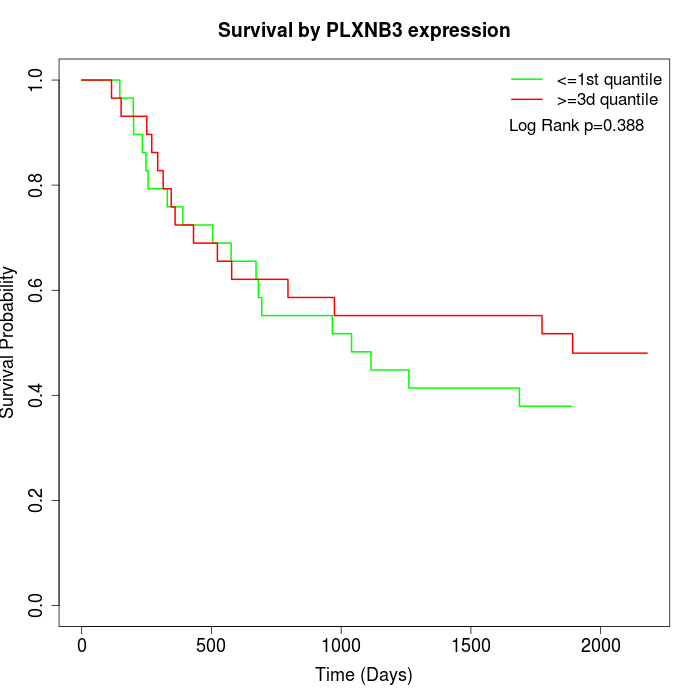
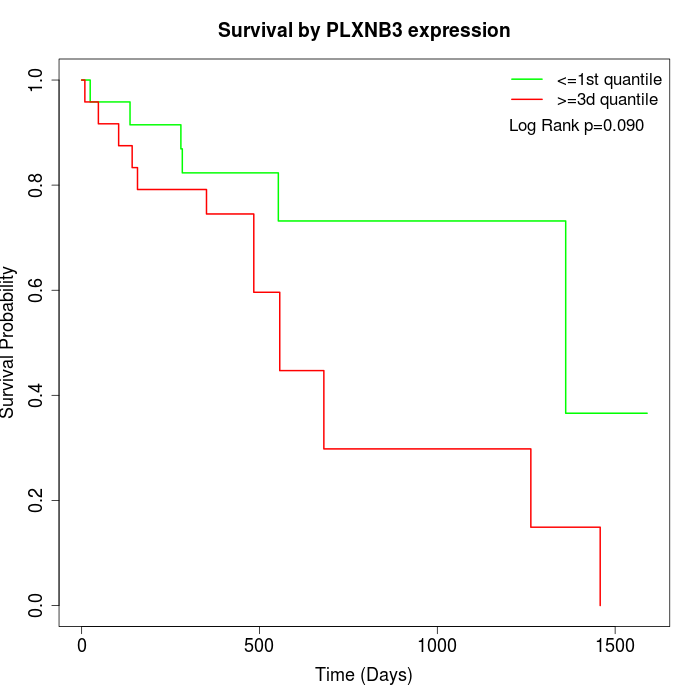
Survival by PLXNB3 expression:
Note: Click image to view full size file.
Copy number change of PLXNB3:
No record found for this gene.
Somatic mutations of PLXNB3:
Generating mutation plots.
Highly correlated genes for PLXNB3:
Showing top 20/433 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLXNB3 | SPEG | 0.793758 | 3 | 0 | 3 |
| PLXNB3 | ZNF362 | 0.774261 | 3 | 0 | 3 |
| PLXNB3 | KRTAP10-9 | 0.768208 | 3 | 0 | 3 |
| PLXNB3 | OR2T33 | 0.76549 | 3 | 0 | 3 |
| PLXNB3 | GSX1 | 0.759359 | 3 | 0 | 3 |
| PLXNB3 | TTLL11 | 0.759119 | 3 | 0 | 3 |
| PLXNB3 | KRTAP12-2 | 0.753511 | 3 | 0 | 3 |
| PLXNB3 | VWA5B1 | 0.742533 | 3 | 0 | 3 |
| PLXNB3 | PDK2 | 0.739658 | 3 | 0 | 3 |
| PLXNB3 | OR1L6 | 0.739576 | 3 | 0 | 3 |
| PLXNB3 | PHF2 | 0.737048 | 3 | 0 | 3 |
| PLXNB3 | RXRG | 0.735806 | 4 | 0 | 3 |
| PLXNB3 | SLC26A5 | 0.731649 | 3 | 0 | 3 |
| PLXNB3 | MEF2A | 0.730697 | 3 | 0 | 3 |
| PLXNB3 | CERS2 | 0.725789 | 3 | 0 | 3 |
| PLXNB3 | PXN | 0.723702 | 3 | 0 | 3 |
| PLXNB3 | RIC8B | 0.722208 | 3 | 0 | 3 |
| PLXNB3 | SEMA4C | 0.717309 | 3 | 0 | 3 |
| PLXNB3 | ALG11 | 0.715311 | 3 | 0 | 3 |
| PLXNB3 | KBTBD2 | 0.706874 | 4 | 0 | 4 |
For details and further investigation, click here