| Full name: peroxisome proliferator activated receptor gamma | Alias Symbol: PPARG1|PPARG2|NR1C3|PPARgamma | ||
| Type: protein-coding gene | Cytoband: 3p25.2 | ||
| Entrez ID: 5468 | HGNC ID: HGNC:9236 | Ensembl Gene: ENSG00000132170 | OMIM ID: 601487 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PPARG involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway | |
| hsa04152 | AMPK signaling pathway | |
| hsa04211 | Longevity regulating pathway - mammal |
Expression of PPARG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPARG | 5468 | 208510_s_at | 0.1445 | 0.9264 | |
| GSE20347 | PPARG | 5468 | 208510_s_at | -0.4140 | 0.0048 | |
| GSE23400 | PPARG | 5468 | 208510_s_at | -0.1424 | 0.0032 | |
| GSE26886 | PPARG | 5468 | 208510_s_at | -0.2525 | 0.4620 | |
| GSE29001 | PPARG | 5468 | 208510_s_at | -0.4630 | 0.0535 | |
| GSE38129 | PPARG | 5468 | 208510_s_at | -0.4855 | 0.0394 | |
| GSE45670 | PPARG | 5468 | 208510_s_at | -0.4954 | 0.1177 | |
| GSE53622 | PPARG | 5468 | 51202 | -1.3499 | 0.0000 | |
| GSE53624 | PPARG | 5468 | 13028 | -1.4369 | 0.0000 | |
| GSE63941 | PPARG | 5468 | 208510_s_at | -2.1877 | 0.0340 | |
| GSE77861 | PPARG | 5468 | 208510_s_at | 0.0997 | 0.6372 | |
| GSE97050 | PPARG | 5468 | A_33_P3350726 | -0.9446 | 0.1958 | |
| SRP007169 | PPARG | 5468 | RNAseq | 0.1113 | 0.8214 | |
| SRP008496 | PPARG | 5468 | RNAseq | -0.4626 | 0.3487 | |
| SRP064894 | PPARG | 5468 | RNAseq | 0.1646 | 0.6855 | |
| SRP133303 | PPARG | 5468 | RNAseq | -1.0322 | 0.0000 | |
| SRP159526 | PPARG | 5468 | RNAseq | 0.0268 | 0.9669 | |
| SRP193095 | PPARG | 5468 | RNAseq | -1.6544 | 0.0000 | |
| SRP219564 | PPARG | 5468 | RNAseq | -0.5539 | 0.3665 | |
| TCGA | PPARG | 5468 | RNAseq | -0.9569 | 0.0009 |
Upregulated datasets: 0; Downregulated datasets: 5.
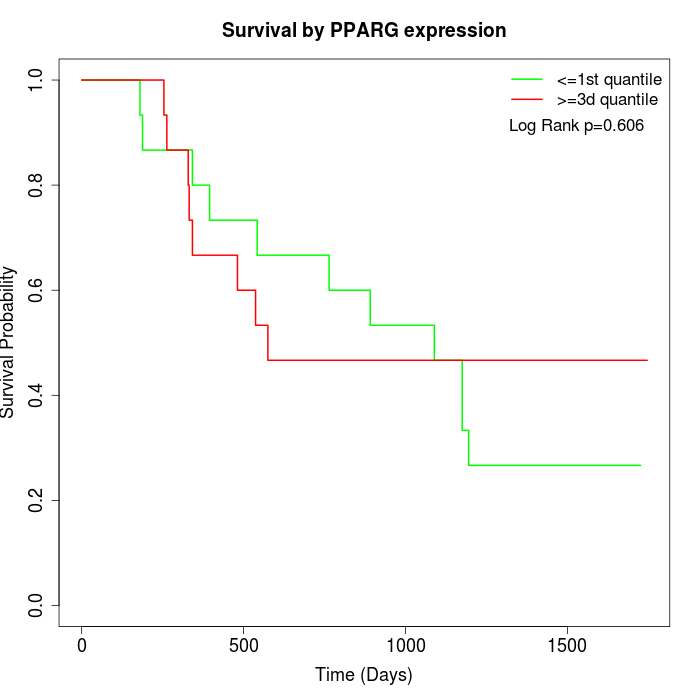
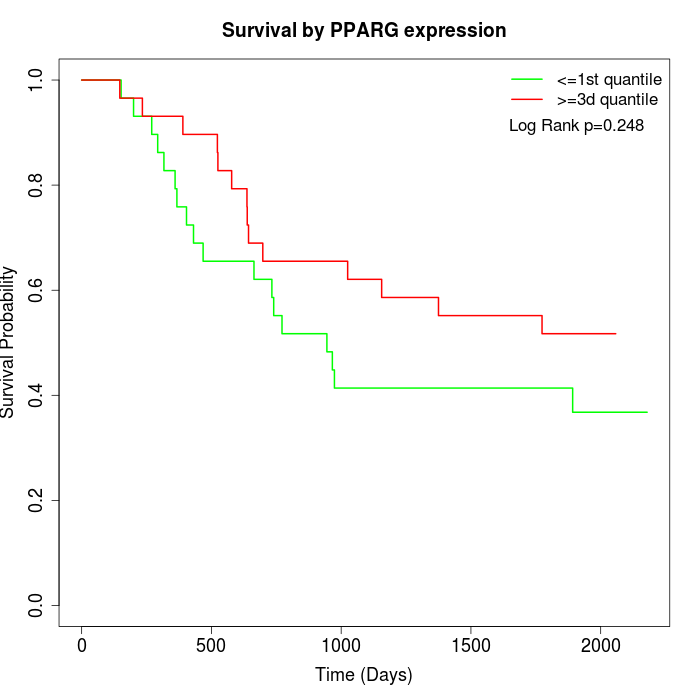
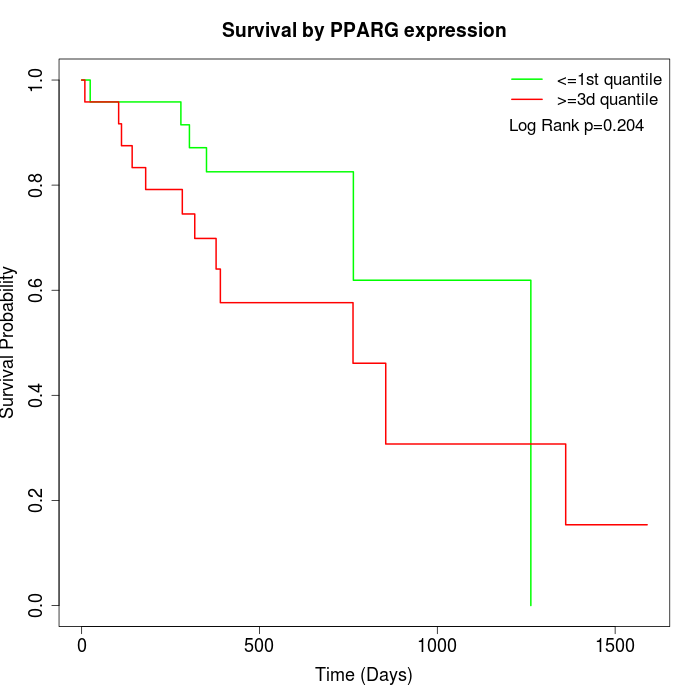
Survival by PPARG expression:
Note: Click image to view full size file.
Copy number change of PPARG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPARG | 5468 | 1 | 17 | 12 | |
| GSE20123 | PPARG | 5468 | 1 | 18 | 11 | |
| GSE43470 | PPARG | 5468 | 0 | 21 | 22 | |
| GSE46452 | PPARG | 5468 | 2 | 17 | 40 | |
| GSE47630 | PPARG | 5468 | 2 | 23 | 15 | |
| GSE54993 | PPARG | 5468 | 6 | 1 | 63 | |
| GSE54994 | PPARG | 5468 | 1 | 33 | 19 | |
| GSE60625 | PPARG | 5468 | 5 | 0 | 6 | |
| GSE74703 | PPARG | 5468 | 0 | 17 | 19 | |
| GSE74704 | PPARG | 5468 | 1 | 10 | 9 | |
| TCGA | PPARG | 5468 | 1 | 68 | 27 |
Total number of gains: 20; Total number of losses: 225; Total Number of normals: 243.
Somatic mutations of PPARG:
Generating mutation plots.
Highly correlated genes for PPARG:
Showing top 20/366 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPARG | MRI1 | 0.799004 | 3 | 0 | 3 |
| PPARG | HIC2 | 0.773657 | 3 | 0 | 3 |
| PPARG | ZNF470 | 0.731754 | 3 | 0 | 3 |
| PPARG | CLU | 0.726552 | 3 | 0 | 3 |
| PPARG | ZFX | 0.723096 | 3 | 0 | 3 |
| PPARG | SHPRH | 0.718495 | 3 | 0 | 3 |
| PPARG | ZNF121 | 0.717077 | 3 | 0 | 3 |
| PPARG | APEX2 | 0.704565 | 3 | 0 | 3 |
| PPARG | LTV1 | 0.704143 | 3 | 0 | 3 |
| PPARG | PEPD | 0.698058 | 4 | 0 | 3 |
| PPARG | RPS19BP1 | 0.693619 | 3 | 0 | 3 |
| PPARG | GYG2 | 0.691747 | 4 | 0 | 3 |
| PPARG | TOMM5 | 0.690957 | 3 | 0 | 3 |
| PPARG | PINK1 | 0.687745 | 3 | 0 | 3 |
| PPARG | FLYWCH2 | 0.682774 | 3 | 0 | 3 |
| PPARG | SRSF6 | 0.681798 | 3 | 0 | 3 |
| PPARG | PROSER1 | 0.675811 | 3 | 0 | 3 |
| PPARG | ZNF35 | 0.665801 | 3 | 0 | 3 |
| PPARG | SLC9A2 | 0.665299 | 3 | 0 | 3 |
| PPARG | LRRC34 | 0.664726 | 3 | 0 | 3 |
For details and further investigation, click here