| Full name: cerebellin 1 precursor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q12.1 | ||
| Entrez ID: 869 | HGNC ID: HGNC:1543 | Ensembl Gene: ENSG00000102924 | OMIM ID: 600432 |
| Drug and gene relationship at DGIdb | |||
Expression of CBLN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CBLN1 | 869 | 205747_at | -0.5979 | 0.0530 | |
| GSE20347 | CBLN1 | 869 | 205747_at | -0.0233 | 0.7629 | |
| GSE23400 | CBLN1 | 869 | 205747_at | -0.1430 | 0.0007 | |
| GSE26886 | CBLN1 | 869 | 205747_at | -0.1105 | 0.2486 | |
| GSE29001 | CBLN1 | 869 | 205747_at | -0.1936 | 0.3656 | |
| GSE38129 | CBLN1 | 869 | 205747_at | -0.0463 | 0.4575 | |
| GSE45670 | CBLN1 | 869 | 205747_at | -0.0674 | 0.5956 | |
| GSE53622 | CBLN1 | 869 | 55667 | -1.5112 | 0.0000 | |
| GSE53624 | CBLN1 | 869 | 55667 | -1.2906 | 0.0000 | |
| GSE63941 | CBLN1 | 869 | 205747_at | 0.1954 | 0.5635 | |
| GSE77861 | CBLN1 | 869 | 205747_at | -0.1248 | 0.2723 | |
| TCGA | CBLN1 | 869 | RNAseq | -1.8405 | 0.0048 |
Upregulated datasets: 0; Downregulated datasets: 3.
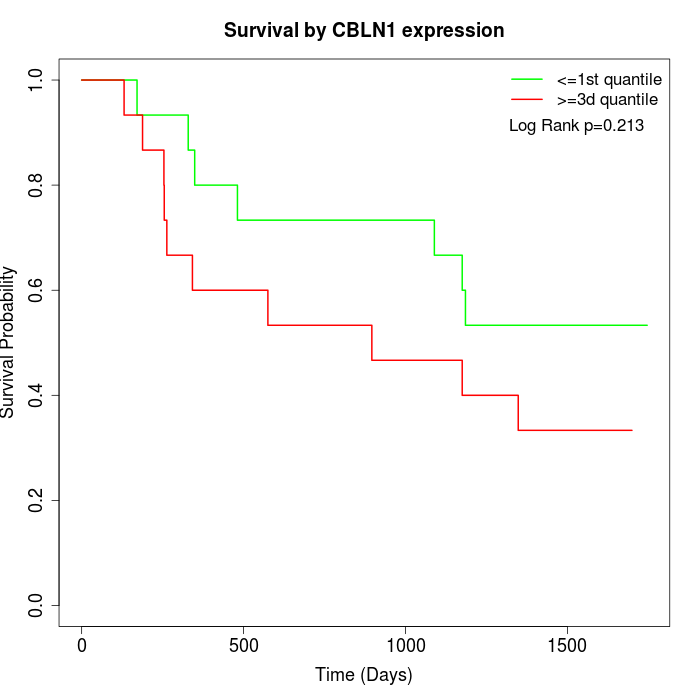
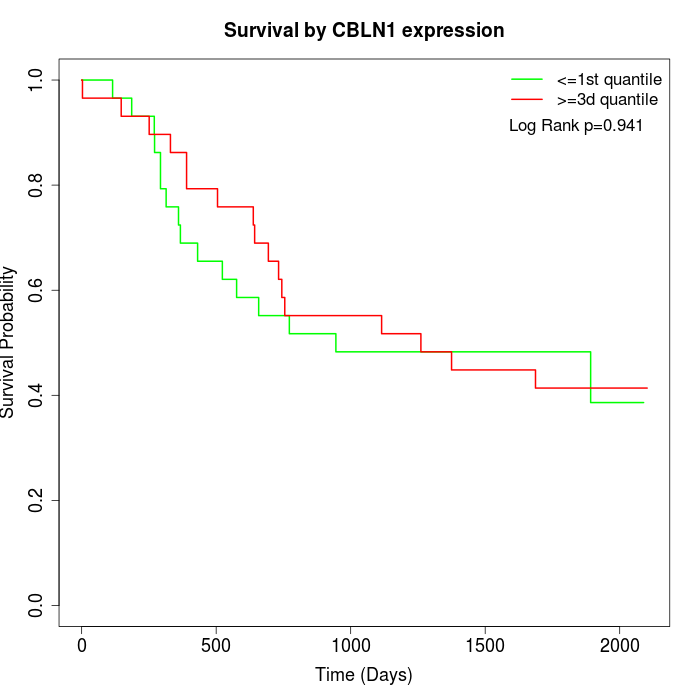
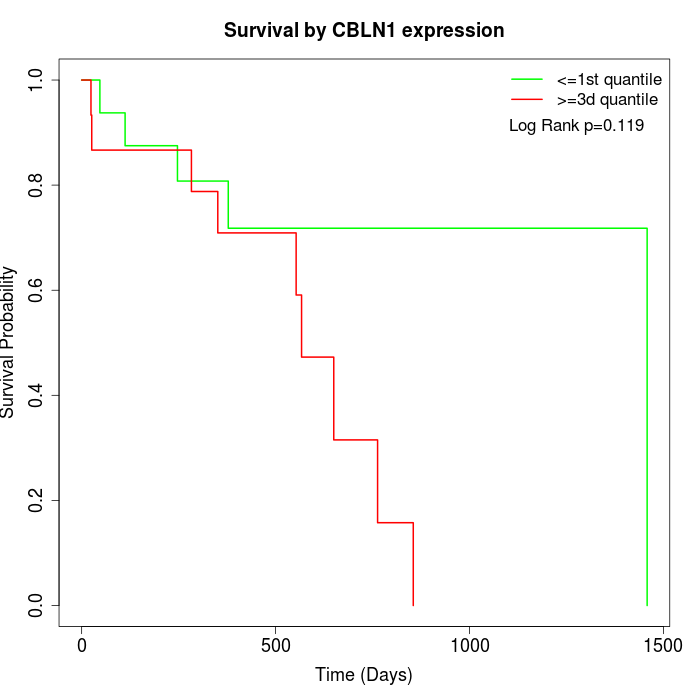
Survival by CBLN1 expression:
Note: Click image to view full size file.
Copy number change of CBLN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CBLN1 | 869 | 4 | 1 | 25 | |
| GSE20123 | CBLN1 | 869 | 3 | 1 | 26 | |
| GSE43470 | CBLN1 | 869 | 2 | 7 | 34 | |
| GSE46452 | CBLN1 | 869 | 38 | 1 | 20 | |
| GSE47630 | CBLN1 | 869 | 11 | 7 | 22 | |
| GSE54993 | CBLN1 | 869 | 2 | 5 | 63 | |
| GSE54994 | CBLN1 | 869 | 6 | 10 | 37 | |
| GSE60625 | CBLN1 | 869 | 4 | 0 | 7 | |
| GSE74703 | CBLN1 | 869 | 2 | 4 | 30 | |
| GSE74704 | CBLN1 | 869 | 3 | 1 | 16 | |
| TCGA | CBLN1 | 869 | 27 | 12 | 57 |
Total number of gains: 102; Total number of losses: 49; Total Number of normals: 337.
Somatic mutations of CBLN1:
Generating mutation plots.
Highly correlated genes for CBLN1:
Showing top 20/788 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CBLN1 | EDDM3A | 0.809276 | 4 | 0 | 4 |
| CBLN1 | LRTM1 | 0.762935 | 4 | 0 | 4 |
| CBLN1 | IFNA5 | 0.739819 | 3 | 0 | 3 |
| CBLN1 | SLC5A4 | 0.731295 | 3 | 0 | 3 |
| CBLN1 | NNAT | 0.718477 | 5 | 0 | 5 |
| CBLN1 | PRB4 | 0.709326 | 5 | 0 | 5 |
| CBLN1 | GDF9 | 0.707068 | 4 | 0 | 4 |
| CBLN1 | CIITA | 0.705809 | 6 | 0 | 6 |
| CBLN1 | GPC5 | 0.702771 | 4 | 0 | 4 |
| CBLN1 | RRH | 0.70164 | 3 | 0 | 3 |
| CBLN1 | CYLC1 | 0.701366 | 4 | 0 | 4 |
| CBLN1 | RORC | 0.700595 | 5 | 0 | 5 |
| CBLN1 | TEK | 0.696191 | 6 | 0 | 4 |
| CBLN1 | KNG1 | 0.69595 | 4 | 0 | 4 |
| CBLN1 | KAT7 | 0.691809 | 3 | 0 | 3 |
| CBLN1 | KCNJ10 | 0.691717 | 4 | 0 | 4 |
| CBLN1 | ACRV1 | 0.688994 | 5 | 0 | 4 |
| CBLN1 | DENND6B | 0.688384 | 3 | 0 | 3 |
| CBLN1 | ODF1 | 0.685402 | 4 | 0 | 3 |
| CBLN1 | GABRA1 | 0.682456 | 4 | 0 | 4 |
For details and further investigation, click here