| Full name: protamine 1 | Alias Symbol: CT94.1 | ||
| Type: protein-coding gene | Cytoband: 16p13.13 | ||
| Entrez ID: 5619 | HGNC ID: HGNC:9447 | Ensembl Gene: ENSG00000175646 | OMIM ID: 182880 |
| Drug and gene relationship at DGIdb | |||
Expression of PRM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRM1 | 5619 | 206358_at | -0.1604 | 0.3839 | |
| GSE20347 | PRM1 | 5619 | 206358_at | -0.0618 | 0.4452 | |
| GSE23400 | PRM1 | 5619 | 206358_at | -0.1422 | 0.0000 | |
| GSE26886 | PRM1 | 5619 | 206358_at | -0.1100 | 0.3331 | |
| GSE29001 | PRM1 | 5619 | 206358_at | -0.1095 | 0.3278 | |
| GSE38129 | PRM1 | 5619 | 206358_at | -0.1243 | 0.0742 | |
| GSE45670 | PRM1 | 5619 | 206358_at | -0.0513 | 0.5815 | |
| GSE53622 | PRM1 | 5619 | 72783 | -0.4614 | 0.0000 | |
| GSE53624 | PRM1 | 5619 | 72783 | -0.6871 | 0.0000 | |
| GSE63941 | PRM1 | 5619 | 206358_at | 0.4125 | 0.0008 | |
| GSE77861 | PRM1 | 5619 | 206358_at | -0.1045 | 0.3531 |
Upregulated datasets: 0; Downregulated datasets: 0.
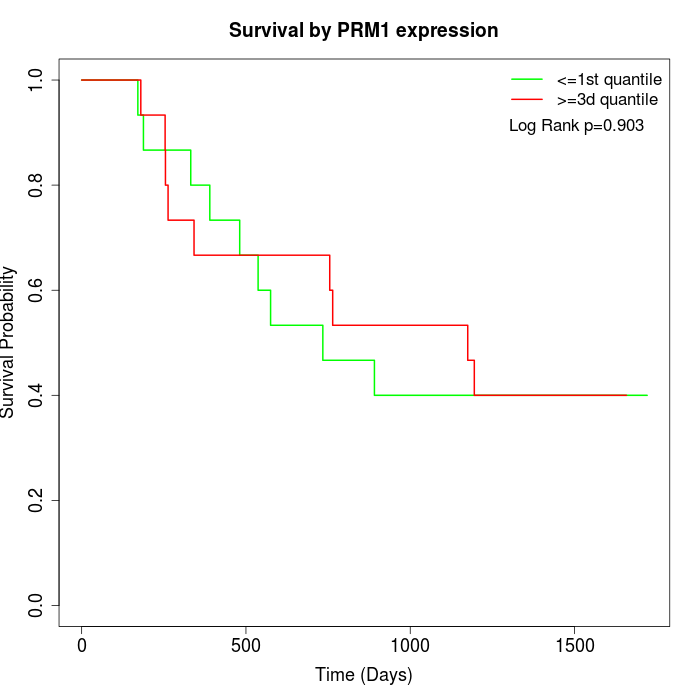
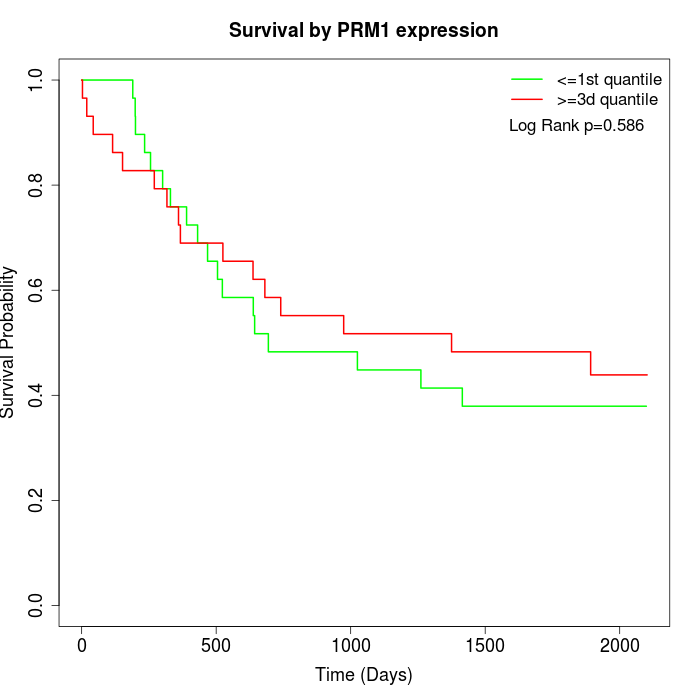
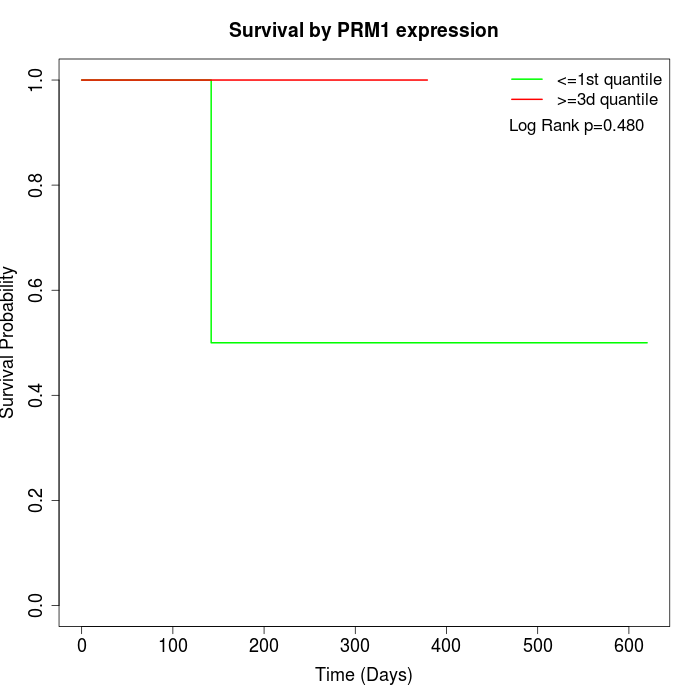
Survival by PRM1 expression:
Note: Click image to view full size file.
Copy number change of PRM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRM1 | 5619 | 5 | 4 | 21 | |
| GSE20123 | PRM1 | 5619 | 5 | 3 | 22 | |
| GSE43470 | PRM1 | 5619 | 3 | 3 | 37 | |
| GSE46452 | PRM1 | 5619 | 38 | 1 | 20 | |
| GSE47630 | PRM1 | 5619 | 14 | 6 | 20 | |
| GSE54993 | PRM1 | 5619 | 3 | 5 | 62 | |
| GSE54994 | PRM1 | 5619 | 5 | 9 | 39 | |
| GSE60625 | PRM1 | 5619 | 4 | 0 | 7 | |
| GSE74703 | PRM1 | 5619 | 3 | 2 | 31 | |
| GSE74704 | PRM1 | 5619 | 2 | 1 | 17 | |
| TCGA | PRM1 | 5619 | 20 | 12 | 64 |
Total number of gains: 102; Total number of losses: 46; Total Number of normals: 340.
Somatic mutations of PRM1:
Generating mutation plots.
Highly correlated genes for PRM1:
Showing top 20/978 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRM1 | C10orf90 | 0.712994 | 3 | 0 | 3 |
| PRM1 | SMYD1 | 0.681446 | 4 | 0 | 4 |
| PRM1 | MEFV | 0.681091 | 5 | 0 | 4 |
| PRM1 | ZMYND10 | 0.678855 | 5 | 0 | 4 |
| PRM1 | RECK | 0.678229 | 5 | 0 | 5 |
| PRM1 | OR51I1 | 0.673323 | 4 | 0 | 4 |
| PRM1 | CAMK2B | 0.668634 | 7 | 0 | 7 |
| PRM1 | BARHL1 | 0.664104 | 4 | 0 | 4 |
| PRM1 | AMPD1 | 0.660669 | 3 | 0 | 3 |
| PRM1 | NCAM1 | 0.660354 | 3 | 0 | 3 |
| PRM1 | STXBP6 | 0.659473 | 3 | 0 | 3 |
| PRM1 | CRHR2 | 0.656245 | 5 | 0 | 4 |
| PRM1 | ST18 | 0.655958 | 3 | 0 | 3 |
| PRM1 | CMTM1 | 0.6524 | 3 | 0 | 3 |
| PRM1 | PKD2L1 | 0.651184 | 8 | 0 | 7 |
| PRM1 | AMBN | 0.648143 | 6 | 0 | 6 |
| PRM1 | MIR4755 | 0.646313 | 4 | 0 | 3 |
| PRM1 | BSND | 0.645913 | 4 | 0 | 4 |
| PRM1 | TEK | 0.640429 | 3 | 0 | 3 |
| PRM1 | TOP3B | 0.640179 | 3 | 0 | 3 |
For details and further investigation, click here