| Full name: PWP2 small subunit processome component | Alias Symbol: EHOC-17|UTP1 | ||
| Type: protein-coding gene | Cytoband: 21q22.3 | ||
| Entrez ID: 5822 | HGNC ID: HGNC:9711 | Ensembl Gene: ENSG00000241945 | OMIM ID: 601475 |
| Drug and gene relationship at DGIdb | |||
Expression of PWP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PWP2 | 5822 | 209336_at | 0.8156 | 0.1332 | |
| GSE20347 | PWP2 | 5822 | 209336_at | 0.3706 | 0.0013 | |
| GSE23400 | PWP2 | 5822 | 209336_at | 0.2716 | 0.0000 | |
| GSE26886 | PWP2 | 5822 | 209336_at | 0.3887 | 0.0602 | |
| GSE29001 | PWP2 | 5822 | 209336_at | 0.5437 | 0.0224 | |
| GSE38129 | PWP2 | 5822 | 209336_at | 0.4587 | 0.0000 | |
| GSE45670 | PWP2 | 5822 | 209336_at | 0.3671 | 0.0095 | |
| GSE63941 | PWP2 | 5822 | 209336_at | -0.1490 | 0.7802 | |
| GSE77861 | PWP2 | 5822 | 1565603_at | -0.1263 | 0.3064 | |
| GSE97050 | PWP2 | 5822 | A_23_P102925 | -0.0885 | 0.6865 | |
| TCGA | PWP2 | 5822 | RNAseq | 0.0954 | 0.1111 |
Upregulated datasets: 0; Downregulated datasets: 0.
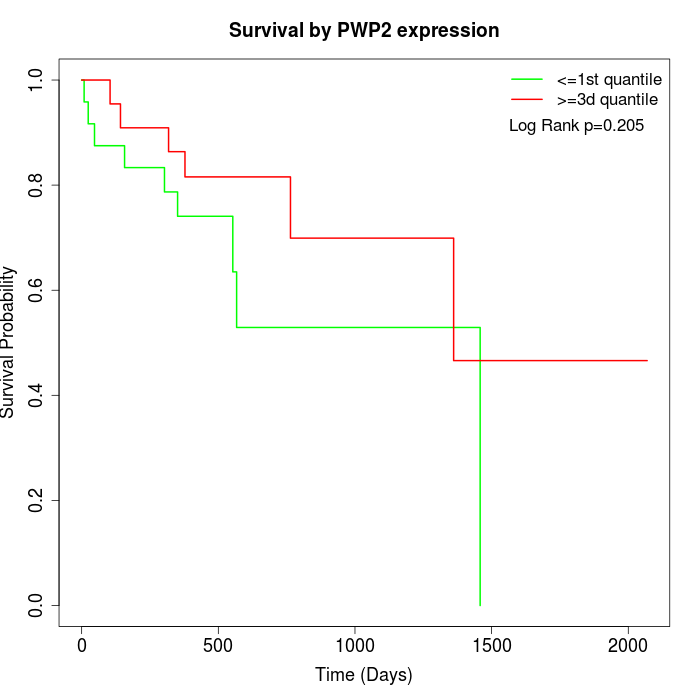
Survival by PWP2 expression:
Note: Click image to view full size file.
Copy number change of PWP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PWP2 | 5822 | 2 | 8 | 20 | |
| GSE20123 | PWP2 | 5822 | 2 | 8 | 20 | |
| GSE43470 | PWP2 | 5822 | 0 | 13 | 30 | |
| GSE46452 | PWP2 | 5822 | 1 | 21 | 37 | |
| GSE47630 | PWP2 | 5822 | 6 | 17 | 17 | |
| GSE54993 | PWP2 | 5822 | 8 | 1 | 61 | |
| GSE54994 | PWP2 | 5822 | 4 | 8 | 41 | |
| GSE60625 | PWP2 | 5822 | 0 | 0 | 11 | |
| GSE74703 | PWP2 | 5822 | 0 | 10 | 26 | |
| GSE74704 | PWP2 | 5822 | 1 | 4 | 15 | |
| TCGA | PWP2 | 5822 | 8 | 39 | 49 |
Total number of gains: 32; Total number of losses: 129; Total Number of normals: 327.
Somatic mutations of PWP2:
Generating mutation plots.
Highly correlated genes for PWP2:
Showing top 20/278 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PWP2 | LRRC73 | 0.727113 | 3 | 0 | 3 |
| PWP2 | ZNF205 | 0.705932 | 3 | 0 | 3 |
| PWP2 | NASP | 0.700196 | 6 | 0 | 6 |
| PWP2 | YIPF2 | 0.696539 | 3 | 0 | 3 |
| PWP2 | RAD18 | 0.690481 | 4 | 0 | 3 |
| PWP2 | PDCD2L | 0.688007 | 3 | 0 | 3 |
| PWP2 | SRM | 0.678242 | 8 | 0 | 8 |
| PWP2 | CCT8 | 0.675405 | 7 | 0 | 6 |
| PWP2 | TRAF7 | 0.656237 | 3 | 0 | 3 |
| PWP2 | ROMO1 | 0.656065 | 3 | 0 | 3 |
| PWP2 | ZNF174 | 0.6528 | 3 | 0 | 3 |
| PWP2 | UBASH3B | 0.648894 | 3 | 0 | 3 |
| PWP2 | MDH2 | 0.648535 | 3 | 0 | 3 |
| PWP2 | ZNF182 | 0.647085 | 3 | 0 | 3 |
| PWP2 | RBM28 | 0.641546 | 8 | 0 | 8 |
| PWP2 | PTBP1 | 0.633662 | 6 | 0 | 5 |
| PWP2 | ZSCAN5A | 0.629752 | 4 | 0 | 3 |
| PWP2 | DHX9 | 0.629485 | 7 | 0 | 7 |
| PWP2 | ZNF8 | 0.628678 | 3 | 0 | 3 |
| PWP2 | PTRH1 | 0.62778 | 4 | 0 | 3 |
For details and further investigation, click here