| Full name: glycogen phosphorylase L | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q22.1 | ||
| Entrez ID: 5836 | HGNC ID: HGNC:9725 | Ensembl Gene: ENSG00000100504 | OMIM ID: 613741 |
| Drug and gene relationship at DGIdb | |||
PYGL involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway | |
| hsa04931 | Insulin resistance |
Expression of PYGL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYGL | 5836 | 202990_at | -0.2515 | 0.8865 | |
| GSE20347 | PYGL | 5836 | 202990_at | -0.5591 | 0.1381 | |
| GSE23400 | PYGL | 5836 | 202990_at | 0.3816 | 0.0297 | |
| GSE26886 | PYGL | 5836 | 202990_at | -0.6637 | 0.0881 | |
| GSE29001 | PYGL | 5836 | 202990_at | -0.0893 | 0.9000 | |
| GSE38129 | PYGL | 5836 | 202990_at | -0.1060 | 0.7825 | |
| GSE45670 | PYGL | 5836 | 202990_at | 0.8660 | 0.0154 | |
| GSE53622 | PYGL | 5836 | 59667 | 0.4783 | 0.0083 | |
| GSE53624 | PYGL | 5836 | 59667 | 0.3496 | 0.0050 | |
| GSE63941 | PYGL | 5836 | 202990_at | -0.5005 | 0.7073 | |
| GSE77861 | PYGL | 5836 | 202990_at | 0.1093 | 0.8882 | |
| GSE97050 | PYGL | 5836 | A_23_P48676 | 0.8618 | 0.0820 | |
| SRP007169 | PYGL | 5836 | RNAseq | 0.1873 | 0.7620 | |
| SRP008496 | PYGL | 5836 | RNAseq | 0.0630 | 0.8584 | |
| SRP064894 | PYGL | 5836 | RNAseq | 0.4833 | 0.0615 | |
| SRP133303 | PYGL | 5836 | RNAseq | 0.2077 | 0.5751 | |
| SRP159526 | PYGL | 5836 | RNAseq | -1.1008 | 0.0016 | |
| SRP193095 | PYGL | 5836 | RNAseq | 0.2305 | 0.5232 | |
| SRP219564 | PYGL | 5836 | RNAseq | 0.2085 | 0.7731 | |
| TCGA | PYGL | 5836 | RNAseq | 0.5985 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
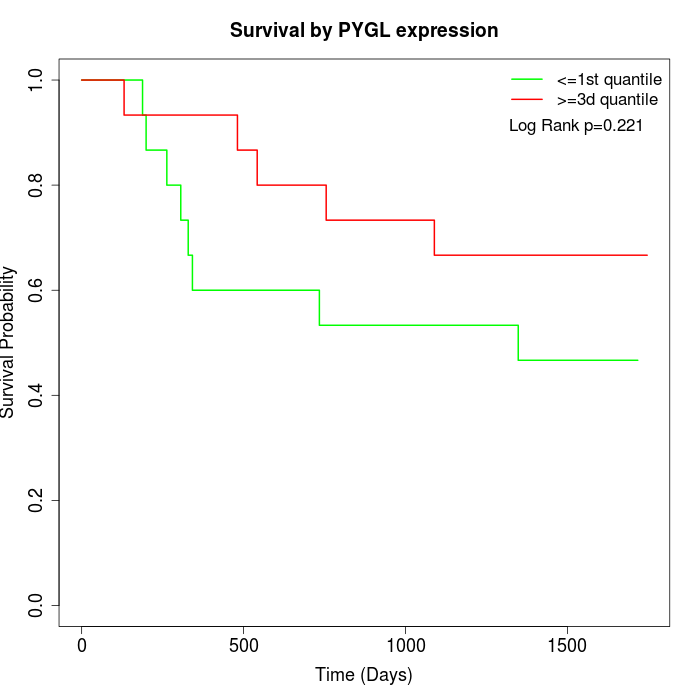
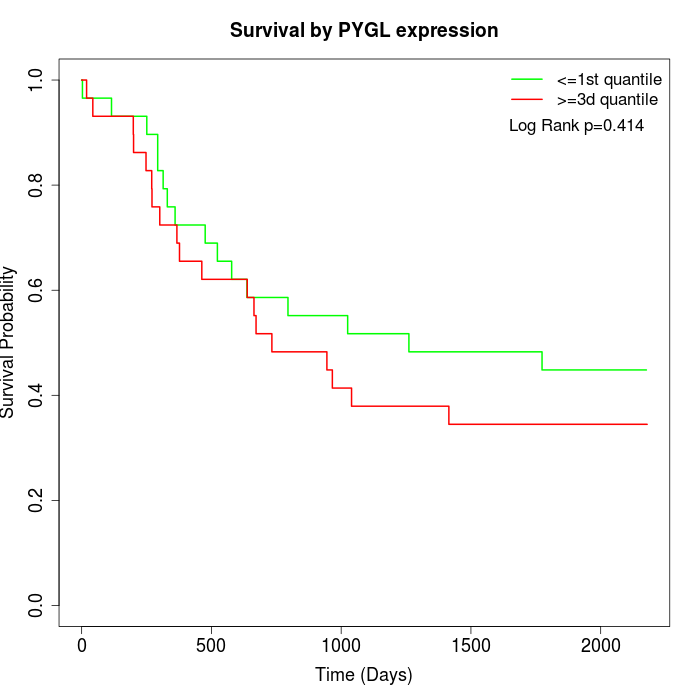
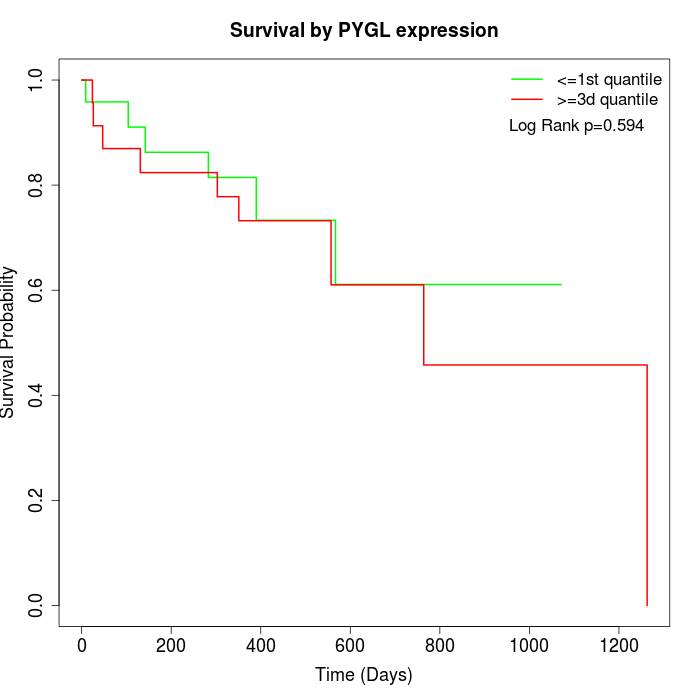
Survival by PYGL expression:
Note: Click image to view full size file.
Copy number change of PYGL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYGL | 5836 | 8 | 3 | 19 | |
| GSE20123 | PYGL | 5836 | 8 | 3 | 19 | |
| GSE43470 | PYGL | 5836 | 9 | 1 | 33 | |
| GSE46452 | PYGL | 5836 | 16 | 3 | 40 | |
| GSE47630 | PYGL | 5836 | 11 | 10 | 19 | |
| GSE54993 | PYGL | 5836 | 3 | 8 | 59 | |
| GSE54994 | PYGL | 5836 | 19 | 4 | 30 | |
| GSE60625 | PYGL | 5836 | 0 | 2 | 9 | |
| GSE74703 | PYGL | 5836 | 8 | 1 | 27 | |
| GSE74704 | PYGL | 5836 | 3 | 2 | 15 | |
| TCGA | PYGL | 5836 | 34 | 13 | 49 |
Total number of gains: 119; Total number of losses: 50; Total Number of normals: 319.
Somatic mutations of PYGL:
Generating mutation plots.
Highly correlated genes for PYGL:
Showing top 20/269 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYGL | GPHN | 0.696893 | 3 | 0 | 3 |
| PYGL | FAM83B | 0.68492 | 5 | 0 | 5 |
| PYGL | FAM110A | 0.677985 | 3 | 0 | 3 |
| PYGL | PTBP3 | 0.672007 | 5 | 0 | 5 |
| PYGL | VSNL1 | 0.666683 | 4 | 0 | 3 |
| PYGL | KRT80 | 0.660287 | 3 | 0 | 3 |
| PYGL | MCTP1 | 0.657059 | 3 | 0 | 3 |
| PYGL | SLC38A9 | 0.649301 | 4 | 0 | 4 |
| PYGL | DEPDC4 | 0.649253 | 3 | 0 | 3 |
| PYGL | SEMA4B | 0.644878 | 5 | 0 | 3 |
| PYGL | ARHGEF5 | 0.642539 | 4 | 0 | 3 |
| PYGL | SLC25A43 | 0.633558 | 4 | 0 | 3 |
| PYGL | TNFAIP8 | 0.629417 | 4 | 0 | 3 |
| PYGL | SLC39A9 | 0.623251 | 5 | 0 | 5 |
| PYGL | SH3D21 | 0.62281 | 4 | 0 | 3 |
| PYGL | GINS4 | 0.621847 | 3 | 0 | 3 |
| PYGL | NXT2 | 0.62097 | 4 | 0 | 3 |
| PYGL | AJUBA | 0.617187 | 5 | 0 | 4 |
| PYGL | LPAR5 | 0.610753 | 5 | 0 | 4 |
| PYGL | SLC22A23 | 0.610664 | 5 | 0 | 3 |
For details and further investigation, click here