| Full name: lysophosphatidic acid receptor 5 | Alias Symbol: KPG_010|LPA5 | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 57121 | HGNC ID: HGNC:13307 | Ensembl Gene: ENSG00000184574 | OMIM ID: 606926 |
| Drug and gene relationship at DGIdb | |||
LPAR5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04072 | Phospholipase D signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway |
Expression of LPAR5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LPAR5 | 57121 | 230252_at | -0.9119 | 0.3685 | |
| GSE26886 | LPAR5 | 57121 | 230252_at | -1.4770 | 0.0001 | |
| GSE45670 | LPAR5 | 57121 | 230252_at | 0.0277 | 0.9267 | |
| GSE53622 | LPAR5 | 57121 | 28859 | -0.8255 | 0.0000 | |
| GSE53624 | LPAR5 | 57121 | 28859 | -1.0780 | 0.0000 | |
| GSE63941 | LPAR5 | 57121 | 230252_at | 2.7534 | 0.0061 | |
| GSE77861 | LPAR5 | 57121 | 230252_at | -0.4753 | 0.3799 | |
| GSE97050 | LPAR5 | 57121 | A_23_P204375 | 0.4927 | 0.6284 | |
| SRP007169 | LPAR5 | 57121 | RNAseq | -1.3246 | 0.0001 | |
| SRP008496 | LPAR5 | 57121 | RNAseq | -1.3332 | 0.0000 | |
| SRP064894 | LPAR5 | 57121 | RNAseq | -1.0802 | 0.0000 | |
| SRP133303 | LPAR5 | 57121 | RNAseq | -1.0785 | 0.0000 | |
| SRP159526 | LPAR5 | 57121 | RNAseq | -1.8731 | 0.0033 | |
| SRP193095 | LPAR5 | 57121 | RNAseq | -1.0042 | 0.0000 | |
| SRP219564 | LPAR5 | 57121 | RNAseq | -1.5194 | 0.0422 | |
| TCGA | LPAR5 | 57121 | RNAseq | 0.0152 | 0.9064 |
Upregulated datasets: 1; Downregulated datasets: 9.
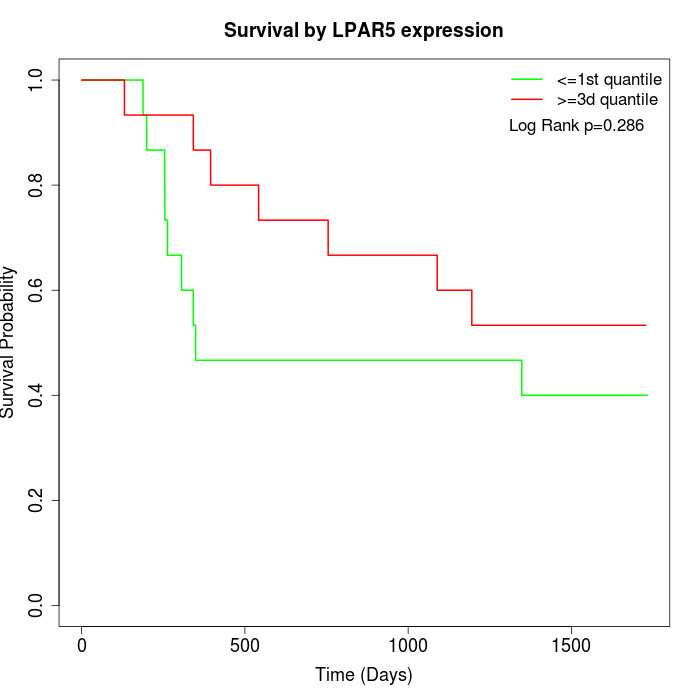
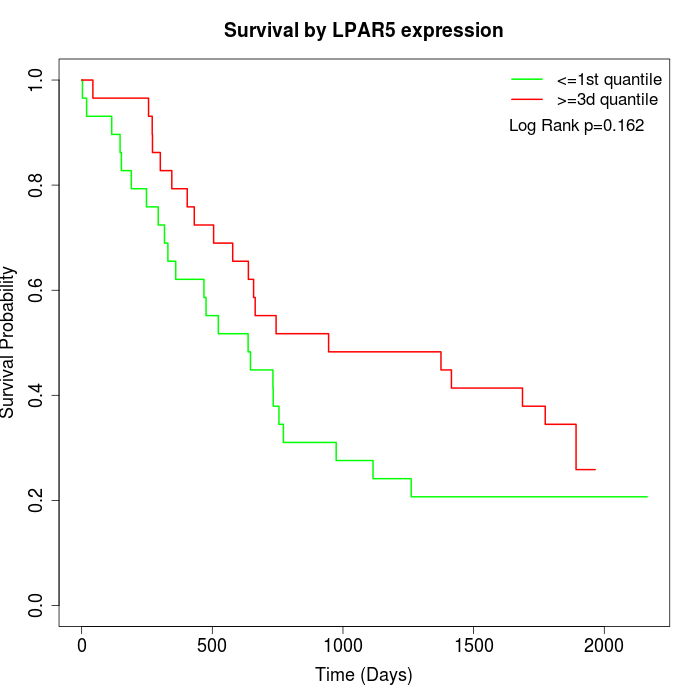
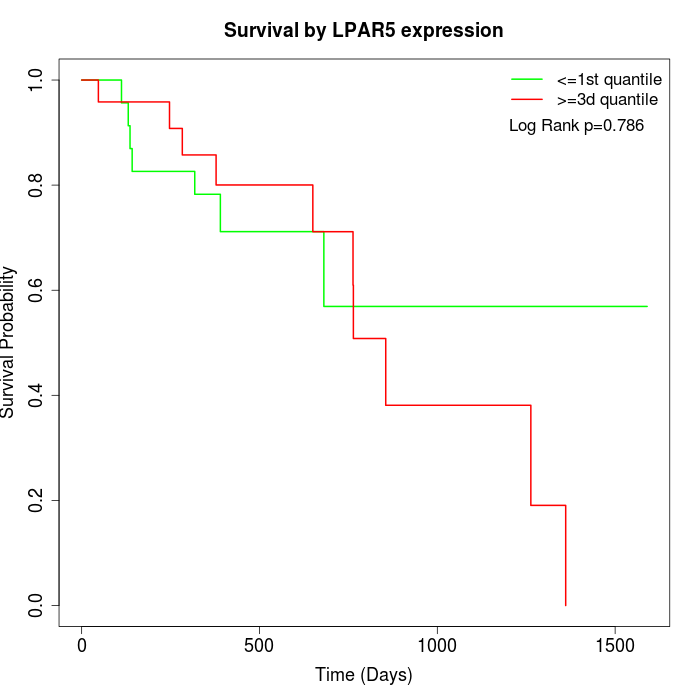
Survival by LPAR5 expression:
Note: Click image to view full size file.
Copy number change of LPAR5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LPAR5 | 57121 | 9 | 4 | 17 | |
| GSE20123 | LPAR5 | 57121 | 9 | 4 | 17 | |
| GSE43470 | LPAR5 | 57121 | 8 | 3 | 32 | |
| GSE46452 | LPAR5 | 57121 | 10 | 1 | 48 | |
| GSE47630 | LPAR5 | 57121 | 12 | 2 | 26 | |
| GSE54993 | LPAR5 | 57121 | 1 | 10 | 59 | |
| GSE54994 | LPAR5 | 57121 | 10 | 2 | 41 | |
| GSE60625 | LPAR5 | 57121 | 0 | 1 | 10 | |
| GSE74703 | LPAR5 | 57121 | 7 | 2 | 27 | |
| GSE74704 | LPAR5 | 57121 | 5 | 3 | 12 | |
| TCGA | LPAR5 | 57121 | 40 | 6 | 50 |
Total number of gains: 111; Total number of losses: 38; Total Number of normals: 339.
Somatic mutations of LPAR5:
Generating mutation plots.
Highly correlated genes for LPAR5:
Showing top 20/891 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LPAR5 | ARAP2 | 0.76214 | 3 | 0 | 3 |
| LPAR5 | PPP1R13L | 0.749956 | 5 | 0 | 5 |
| LPAR5 | CKMT1A | 0.744254 | 3 | 0 | 3 |
| LPAR5 | CERS3 | 0.740091 | 6 | 0 | 6 |
| LPAR5 | DSC2 | 0.73856 | 8 | 0 | 7 |
| LPAR5 | NIPAL1 | 0.730943 | 3 | 0 | 3 |
| LPAR5 | CTNND1 | 0.727865 | 3 | 0 | 3 |
| LPAR5 | DUOXA1 | 0.724296 | 7 | 0 | 6 |
| LPAR5 | KRT75 | 0.719831 | 3 | 0 | 3 |
| LPAR5 | SLC9A3R1 | 0.719256 | 8 | 0 | 7 |
| LPAR5 | NUAK2 | 0.716343 | 3 | 0 | 3 |
| LPAR5 | SERPINB5 | 0.715334 | 8 | 0 | 7 |
| LPAR5 | GJB5 | 0.714999 | 7 | 0 | 7 |
| LPAR5 | CDCP1 | 0.713207 | 5 | 0 | 5 |
| LPAR5 | CDA | 0.710512 | 6 | 0 | 5 |
| LPAR5 | DHRS1 | 0.710476 | 5 | 0 | 5 |
| LPAR5 | ADH7 | 0.708129 | 6 | 0 | 5 |
| LPAR5 | FAM160A1 | 0.707423 | 6 | 0 | 6 |
| LPAR5 | ELOVL1 | 0.702887 | 4 | 0 | 4 |
| LPAR5 | PGLYRP3 | 0.702845 | 4 | 0 | 4 |
For details and further investigation, click here