| Full name: RAB3 GTPase activating non-catalytic protein subunit 2 | Alias Symbol: RAB3-GAP150|KIAA0839|DKFZP434D245|SPG69 | ||
| Type: protein-coding gene | Cytoband: 1q41 | ||
| Entrez ID: 25782 | HGNC ID: HGNC:17168 | Ensembl Gene: ENSG00000118873 | OMIM ID: 609275 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RAB3GAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAB3GAP2 | 25782 | 216057_at | -0.0324 | 0.9494 | |
| GSE20347 | RAB3GAP2 | 25782 | 216057_at | 0.0214 | 0.7526 | |
| GSE23400 | RAB3GAP2 | 25782 | 216057_at | -0.0118 | 0.4902 | |
| GSE26886 | RAB3GAP2 | 25782 | 216057_at | 0.0296 | 0.8322 | |
| GSE29001 | RAB3GAP2 | 25782 | 216057_at | -0.0186 | 0.9182 | |
| GSE38129 | RAB3GAP2 | 25782 | 216057_at | -0.0446 | 0.3783 | |
| GSE45670 | RAB3GAP2 | 25782 | 216057_at | 0.0706 | 0.3510 | |
| GSE53622 | RAB3GAP2 | 25782 | 108118 | -0.2509 | 0.0000 | |
| GSE53624 | RAB3GAP2 | 25782 | 72665 | -0.1033 | 0.0346 | |
| GSE63941 | RAB3GAP2 | 25782 | 216057_at | 0.1100 | 0.5724 | |
| GSE77861 | RAB3GAP2 | 25782 | 216057_at | -0.1306 | 0.1931 | |
| GSE97050 | RAB3GAP2 | 25782 | A_24_P127928 | -0.4385 | 0.1873 | |
| SRP007169 | RAB3GAP2 | 25782 | RNAseq | -0.2750 | 0.4740 | |
| SRP008496 | RAB3GAP2 | 25782 | RNAseq | -0.1836 | 0.3643 | |
| SRP064894 | RAB3GAP2 | 25782 | RNAseq | 0.0169 | 0.9342 | |
| SRP133303 | RAB3GAP2 | 25782 | RNAseq | 0.1517 | 0.2398 | |
| SRP159526 | RAB3GAP2 | 25782 | RNAseq | -0.1149 | 0.7412 | |
| SRP193095 | RAB3GAP2 | 25782 | RNAseq | 0.0418 | 0.5838 | |
| SRP219564 | RAB3GAP2 | 25782 | RNAseq | 0.1397 | 0.6755 | |
| TCGA | RAB3GAP2 | 25782 | RNAseq | 0.0341 | 0.4557 |
Upregulated datasets: 0; Downregulated datasets: 0.
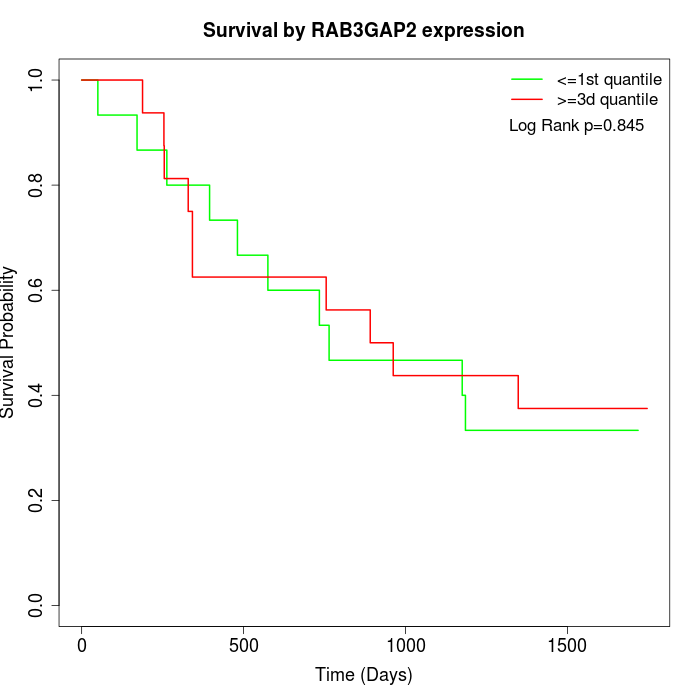
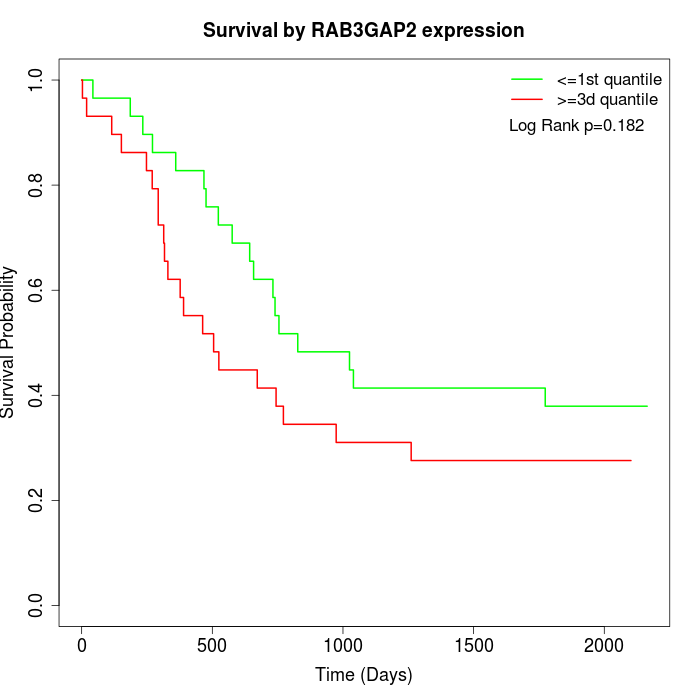
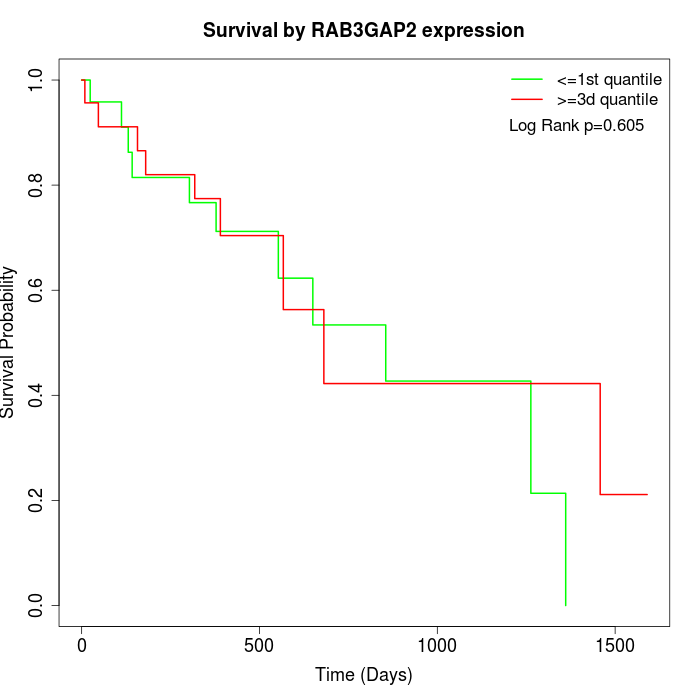
Survival by RAB3GAP2 expression:
Note: Click image to view full size file.
Copy number change of RAB3GAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAB3GAP2 | 25782 | 9 | 0 | 21 | |
| GSE20123 | RAB3GAP2 | 25782 | 9 | 0 | 21 | |
| GSE43470 | RAB3GAP2 | 25782 | 8 | 1 | 34 | |
| GSE46452 | RAB3GAP2 | 25782 | 3 | 1 | 55 | |
| GSE47630 | RAB3GAP2 | 25782 | 15 | 0 | 25 | |
| GSE54993 | RAB3GAP2 | 25782 | 0 | 6 | 64 | |
| GSE54994 | RAB3GAP2 | 25782 | 15 | 0 | 38 | |
| GSE60625 | RAB3GAP2 | 25782 | 0 | 0 | 11 | |
| GSE74703 | RAB3GAP2 | 25782 | 8 | 1 | 27 | |
| GSE74704 | RAB3GAP2 | 25782 | 3 | 0 | 17 | |
| TCGA | RAB3GAP2 | 25782 | 46 | 3 | 47 |
Total number of gains: 116; Total number of losses: 12; Total Number of normals: 360.
Somatic mutations of RAB3GAP2:
Generating mutation plots.
Highly correlated genes for RAB3GAP2:
Showing top 20/60 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAB3GAP2 | FLYWCH2 | 0.7929 | 3 | 0 | 3 |
| RAB3GAP2 | XRRA1 | 0.769545 | 3 | 0 | 3 |
| RAB3GAP2 | TOX4 | 0.697642 | 3 | 0 | 3 |
| RAB3GAP2 | PLD6 | 0.691988 | 3 | 0 | 3 |
| RAB3GAP2 | TPSG1 | 0.680186 | 3 | 0 | 3 |
| RAB3GAP2 | CAPN3 | 0.672374 | 4 | 0 | 3 |
| RAB3GAP2 | CUL3 | 0.670634 | 3 | 0 | 3 |
| RAB3GAP2 | ACOX3 | 0.663997 | 3 | 0 | 3 |
| RAB3GAP2 | CIDEB | 0.658614 | 3 | 0 | 3 |
| RAB3GAP2 | PAQR7 | 0.649115 | 4 | 0 | 3 |
| RAB3GAP2 | SNORD8 | 0.645453 | 4 | 0 | 3 |
| RAB3GAP2 | CCDC97 | 0.641125 | 4 | 0 | 3 |
| RAB3GAP2 | CRYBB2 | 0.635405 | 5 | 0 | 4 |
| RAB3GAP2 | UBE2F | 0.632375 | 4 | 0 | 3 |
| RAB3GAP2 | NOTCH4 | 0.625851 | 5 | 0 | 4 |
| RAB3GAP2 | ZNF214 | 0.624063 | 4 | 0 | 3 |
| RAB3GAP2 | LDB3 | 0.621762 | 4 | 0 | 3 |
| RAB3GAP2 | ARSK | 0.618257 | 3 | 0 | 3 |
| RAB3GAP2 | WDFY3 | 0.613215 | 4 | 0 | 3 |
| RAB3GAP2 | TNP2 | 0.608124 | 5 | 0 | 4 |
For details and further investigation, click here