| Full name: RasGEF domain family member 1C | Alias Symbol: FLJ35841 | ||
| Type: protein-coding gene | Cytoband: 5q35.3 | ||
| Entrez ID: 255426 | HGNC ID: HGNC:27400 | Ensembl Gene: ENSG00000146090 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of RASGEF1C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RASGEF1C | 255426 | 236748_at | -0.0661 | 0.7429 | |
| GSE26886 | RASGEF1C | 255426 | 236748_at | 0.1265 | 0.3943 | |
| GSE45670 | RASGEF1C | 255426 | 236748_at | -0.0410 | 0.6989 | |
| GSE53622 | RASGEF1C | 255426 | 40165 | -0.9612 | 0.0000 | |
| GSE53624 | RASGEF1C | 255426 | 40165 | -0.5255 | 0.0004 | |
| GSE63941 | RASGEF1C | 255426 | 236748_at | 0.1023 | 0.4885 | |
| GSE77861 | RASGEF1C | 255426 | 236748_at | -0.1252 | 0.2326 | |
| GSE97050 | RASGEF1C | 255426 | A_24_P383762 | -0.2902 | 0.4576 | |
| TCGA | RASGEF1C | 255426 | RNAseq | -1.3745 | 0.0274 |
Upregulated datasets: 0; Downregulated datasets: 1.
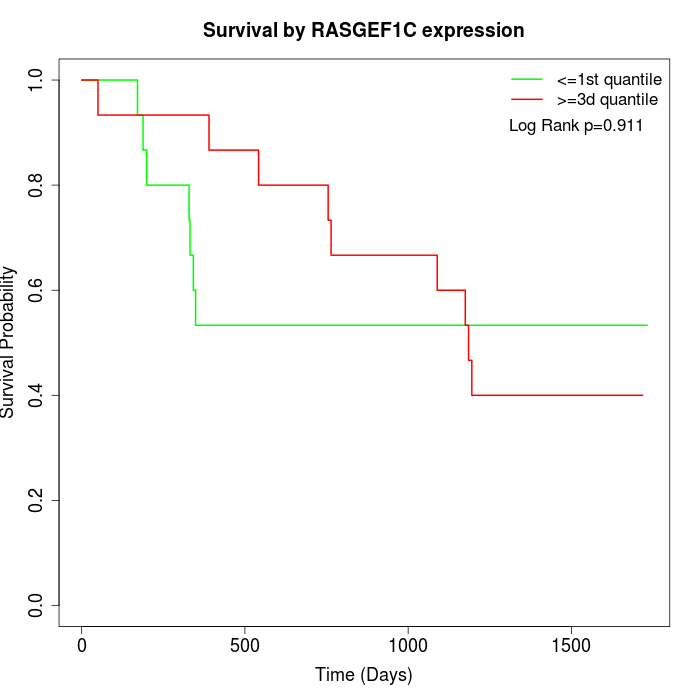
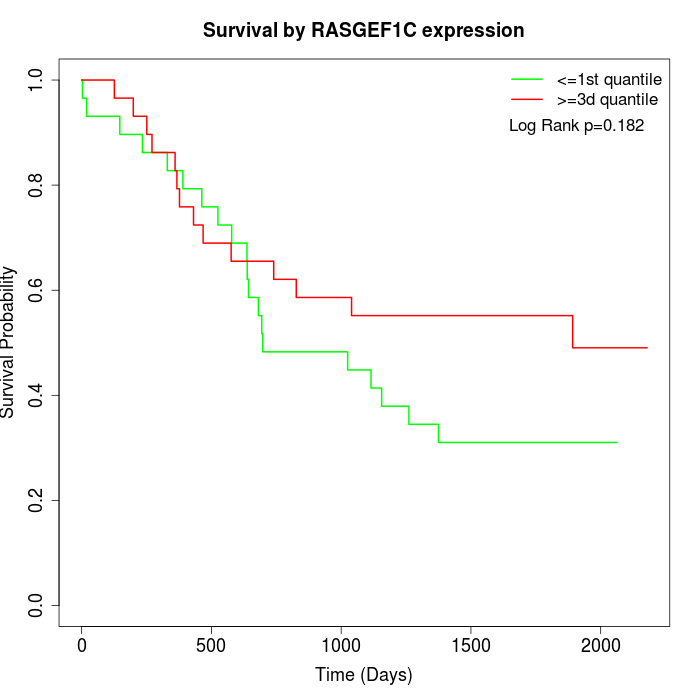
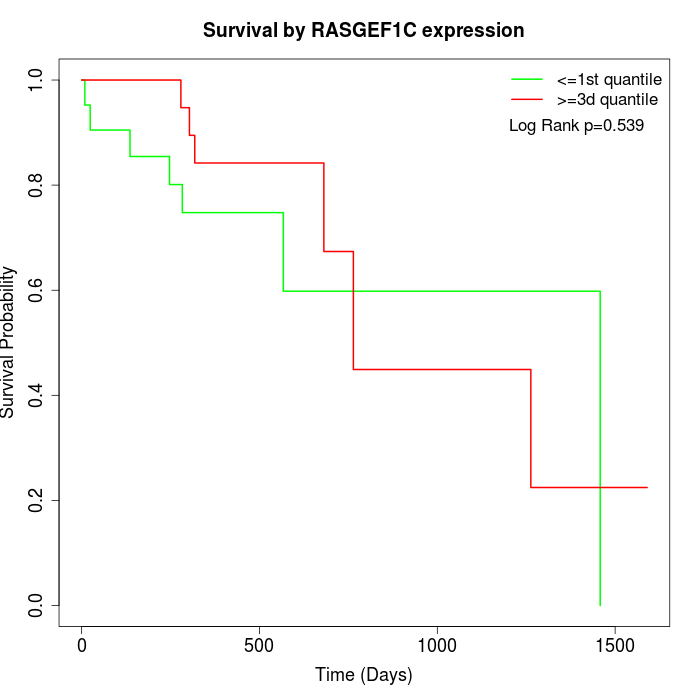
Survival by RASGEF1C expression:
Note: Click image to view full size file.
Copy number change of RASGEF1C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RASGEF1C | 255426 | 2 | 12 | 16 | |
| GSE20123 | RASGEF1C | 255426 | 2 | 12 | 16 | |
| GSE43470 | RASGEF1C | 255426 | 1 | 11 | 31 | |
| GSE46452 | RASGEF1C | 255426 | 0 | 27 | 32 | |
| GSE47630 | RASGEF1C | 255426 | 1 | 20 | 19 | |
| GSE54993 | RASGEF1C | 255426 | 10 | 2 | 58 | |
| GSE54994 | RASGEF1C | 255426 | 3 | 17 | 33 | |
| GSE60625 | RASGEF1C | 255426 | 1 | 0 | 10 | |
| GSE74703 | RASGEF1C | 255426 | 1 | 7 | 28 | |
| GSE74704 | RASGEF1C | 255426 | 1 | 6 | 13 | |
| TCGA | RASGEF1C | 255426 | 12 | 34 | 50 |
Total number of gains: 34; Total number of losses: 148; Total Number of normals: 306.
Somatic mutations of RASGEF1C:
Generating mutation plots.
Highly correlated genes for RASGEF1C:
Showing top 20/128 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RASGEF1C | OPN4 | 0.738953 | 3 | 0 | 3 |
| RASGEF1C | PRSS8 | 0.7232 | 3 | 0 | 3 |
| RASGEF1C | CLDN24 | 0.719268 | 3 | 0 | 3 |
| RASGEF1C | ZNF775 | 0.714851 | 3 | 0 | 3 |
| RASGEF1C | CRYBA2 | 0.689058 | 5 | 0 | 4 |
| RASGEF1C | CA5A | 0.678898 | 3 | 0 | 3 |
| RASGEF1C | DHRS7C | 0.677547 | 3 | 0 | 3 |
| RASGEF1C | DHH | 0.675193 | 3 | 0 | 3 |
| RASGEF1C | TULP1 | 0.669185 | 3 | 0 | 3 |
| RASGEF1C | IQCF3 | 0.665091 | 3 | 0 | 3 |
| RASGEF1C | C16orf89 | 0.659323 | 3 | 0 | 3 |
| RASGEF1C | ACTL6B | 0.65531 | 4 | 0 | 4 |
| RASGEF1C | LDLRAD1 | 0.655159 | 3 | 0 | 3 |
| RASGEF1C | BMP10 | 0.648743 | 3 | 0 | 3 |
| RASGEF1C | NPY2R | 0.644748 | 3 | 0 | 3 |
| RASGEF1C | SRRM3 | 0.642829 | 4 | 0 | 4 |
| RASGEF1C | C20orf173 | 0.642708 | 4 | 0 | 3 |
| RASGEF1C | CRB2 | 0.64244 | 4 | 0 | 3 |
| RASGEF1C | PHF21B | 0.639933 | 4 | 0 | 3 |
| RASGEF1C | PMM1 | 0.636462 | 4 | 0 | 3 |
For details and further investigation, click here