| Full name: phosphomannomutase 1 | Alias Symbol: Sec53 | ||
| Type: protein-coding gene | Cytoband: 22q13.2 | ||
| Entrez ID: 5372 | HGNC ID: HGNC:9114 | Ensembl Gene: ENSG00000100417 | OMIM ID: 601786 |
| Drug and gene relationship at DGIdb | |||
Expression of PMM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PMM1 | 5372 | 203467_at | -1.3271 | 0.0498 | |
| GSE20347 | PMM1 | 5372 | 203467_at | -1.7303 | 0.0000 | |
| GSE23400 | PMM1 | 5372 | 203467_at | -1.2766 | 0.0000 | |
| GSE26886 | PMM1 | 5372 | 203467_at | -1.6263 | 0.0000 | |
| GSE29001 | PMM1 | 5372 | 203467_at | -1.6797 | 0.0000 | |
| GSE38129 | PMM1 | 5372 | 203467_at | -1.4498 | 0.0000 | |
| GSE45670 | PMM1 | 5372 | 203467_at | -1.2428 | 0.0000 | |
| GSE53622 | PMM1 | 5372 | 7493 | -1.4900 | 0.0000 | |
| GSE53624 | PMM1 | 5372 | 7493 | -1.7313 | 0.0000 | |
| GSE63941 | PMM1 | 5372 | 203467_at | -1.5533 | 0.0118 | |
| GSE77861 | PMM1 | 5372 | 203467_at | -0.9503 | 0.0133 | |
| GSE97050 | PMM1 | 5372 | A_23_P211598 | -0.4606 | 0.3240 | |
| SRP007169 | PMM1 | 5372 | RNAseq | -3.7746 | 0.0000 | |
| SRP008496 | PMM1 | 5372 | RNAseq | -3.5415 | 0.0000 | |
| SRP064894 | PMM1 | 5372 | RNAseq | -1.2571 | 0.0000 | |
| SRP133303 | PMM1 | 5372 | RNAseq | -1.5514 | 0.0000 | |
| SRP159526 | PMM1 | 5372 | RNAseq | -1.7448 | 0.0000 | |
| SRP193095 | PMM1 | 5372 | RNAseq | -1.5132 | 0.0000 | |
| SRP219564 | PMM1 | 5372 | RNAseq | -1.7012 | 0.0019 | |
| TCGA | PMM1 | 5372 | RNAseq | -0.3390 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 17.
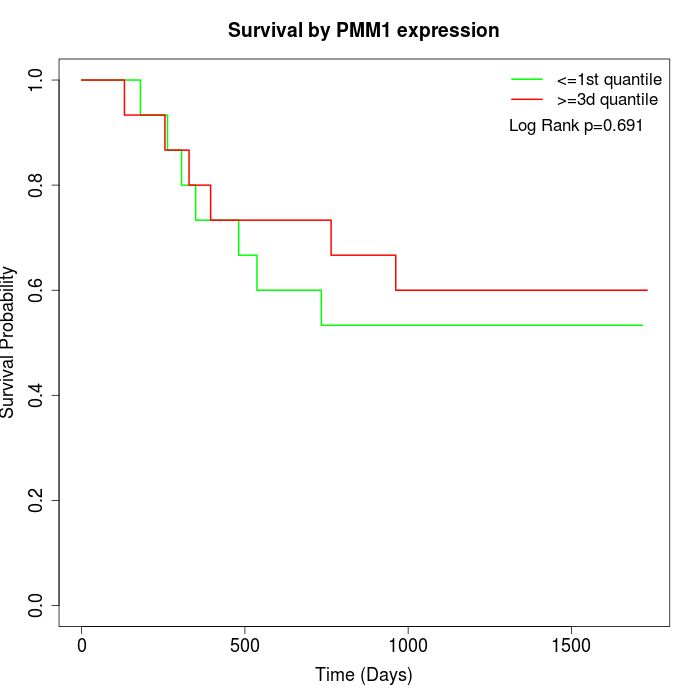
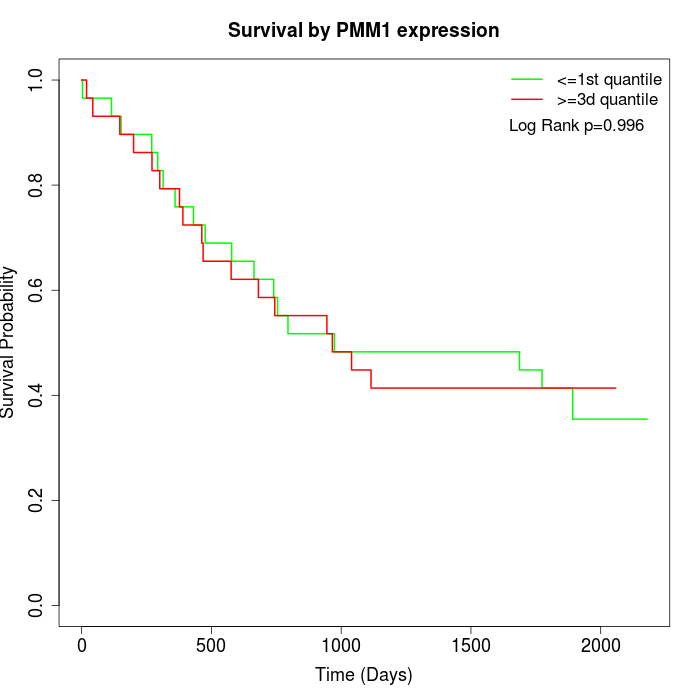
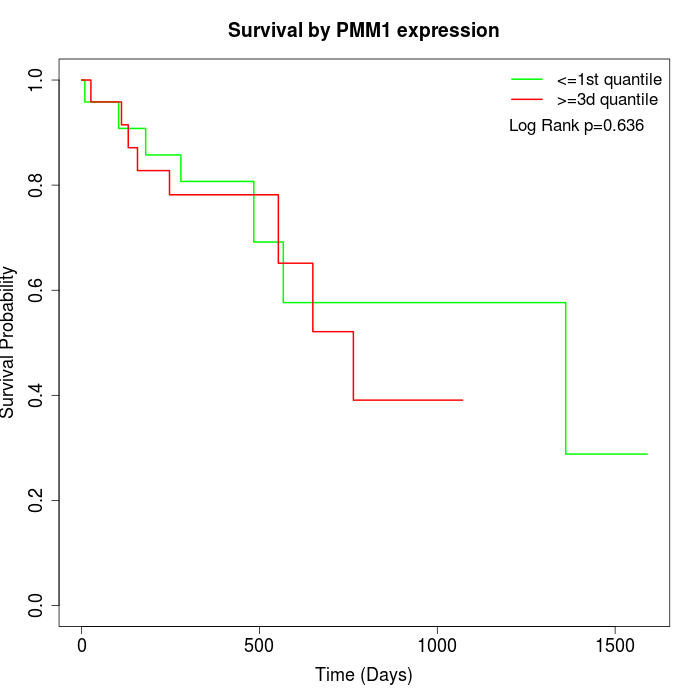
Survival by PMM1 expression:
Note: Click image to view full size file.
Copy number change of PMM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PMM1 | 5372 | 6 | 5 | 19 | |
| GSE20123 | PMM1 | 5372 | 6 | 4 | 20 | |
| GSE43470 | PMM1 | 5372 | 5 | 6 | 32 | |
| GSE46452 | PMM1 | 5372 | 31 | 2 | 26 | |
| GSE47630 | PMM1 | 5372 | 9 | 4 | 27 | |
| GSE54993 | PMM1 | 5372 | 3 | 6 | 61 | |
| GSE54994 | PMM1 | 5372 | 11 | 8 | 34 | |
| GSE60625 | PMM1 | 5372 | 5 | 0 | 6 | |
| GSE74703 | PMM1 | 5372 | 5 | 4 | 27 | |
| GSE74704 | PMM1 | 5372 | 3 | 2 | 15 | |
| TCGA | PMM1 | 5372 | 26 | 14 | 56 |
Total number of gains: 110; Total number of losses: 55; Total Number of normals: 323.
Somatic mutations of PMM1:
Generating mutation plots.
Highly correlated genes for PMM1:
Showing top 20/829 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PMM1 | GYS2 | 0.869167 | 10 | 0 | 10 |
| PMM1 | PAQR8 | 0.857493 | 6 | 0 | 6 |
| PMM1 | FAM163B | 0.851933 | 3 | 0 | 3 |
| PMM1 | RANBP9 | 0.849414 | 10 | 0 | 10 |
| PMM1 | MUC22 | 0.848391 | 3 | 0 | 3 |
| PMM1 | UPK1A | 0.846181 | 11 | 0 | 11 |
| PMM1 | ACER1 | 0.843918 | 7 | 0 | 7 |
| PMM1 | NUCB2 | 0.843299 | 11 | 0 | 11 |
| PMM1 | SCNN1B | 0.838705 | 11 | 0 | 11 |
| PMM1 | SAMD5 | 0.835497 | 6 | 0 | 6 |
| PMM1 | CRISP3 | 0.835473 | 10 | 0 | 10 |
| PMM1 | UBL3 | 0.835413 | 11 | 0 | 11 |
| PMM1 | VAT1 | 0.835137 | 11 | 0 | 11 |
| PMM1 | ST3GAL4 | 0.833644 | 11 | 0 | 11 |
| PMM1 | GDPD3 | 0.833359 | 11 | 0 | 11 |
| PMM1 | ECM1 | 0.832496 | 12 | 0 | 12 |
| PMM1 | SH3BGRL2 | 0.829557 | 6 | 0 | 6 |
| PMM1 | MXD1 | 0.827488 | 11 | 0 | 11 |
| PMM1 | DUSP5 | 0.827111 | 10 | 0 | 10 |
| PMM1 | EHD3 | 0.826761 | 11 | 0 | 11 |
For details and further investigation, click here