| Full name: regulator of G protein signaling 12 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4p16.3 | ||
| Entrez ID: 6002 | HGNC ID: HGNC:9994 | Ensembl Gene: ENSG00000159788 | OMIM ID: 602512 |
| Drug and gene relationship at DGIdb | |||
Expression of RGS12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS12 | 6002 | 205823_at | 0.0715 | 0.9431 | |
| GSE20347 | RGS12 | 6002 | 205823_at | -0.8939 | 0.0000 | |
| GSE23400 | RGS12 | 6002 | 205823_at | -0.6613 | 0.0000 | |
| GSE26886 | RGS12 | 6002 | 205823_at | -1.2062 | 0.0000 | |
| GSE29001 | RGS12 | 6002 | 205823_at | -0.7092 | 0.0384 | |
| GSE38129 | RGS12 | 6002 | 205823_at | -0.5572 | 0.0130 | |
| GSE45670 | RGS12 | 6002 | 242079_at | 0.0218 | 0.9052 | |
| GSE53622 | RGS12 | 6002 | 47634 | -0.4244 | 0.0000 | |
| GSE53624 | RGS12 | 6002 | 77195 | -0.7821 | 0.0000 | |
| GSE63941 | RGS12 | 6002 | 205823_at | -0.2392 | 0.4876 | |
| GSE77861 | RGS12 | 6002 | 205823_at | -0.4224 | 0.0814 | |
| GSE97050 | RGS12 | 6002 | A_23_P352435 | -0.1392 | 0.5880 | |
| SRP007169 | RGS12 | 6002 | RNAseq | -1.0258 | 0.0403 | |
| SRP008496 | RGS12 | 6002 | RNAseq | -0.9932 | 0.0029 | |
| SRP064894 | RGS12 | 6002 | RNAseq | -0.8509 | 0.0000 | |
| SRP133303 | RGS12 | 6002 | RNAseq | -0.9070 | 0.0000 | |
| SRP159526 | RGS12 | 6002 | RNAseq | -0.6870 | 0.1935 | |
| SRP193095 | RGS12 | 6002 | RNAseq | -0.8046 | 0.0001 | |
| SRP219564 | RGS12 | 6002 | RNAseq | -0.7047 | 0.1728 | |
| TCGA | RGS12 | 6002 | RNAseq | 0.0837 | 0.2673 |
Upregulated datasets: 0; Downregulated datasets: 2.
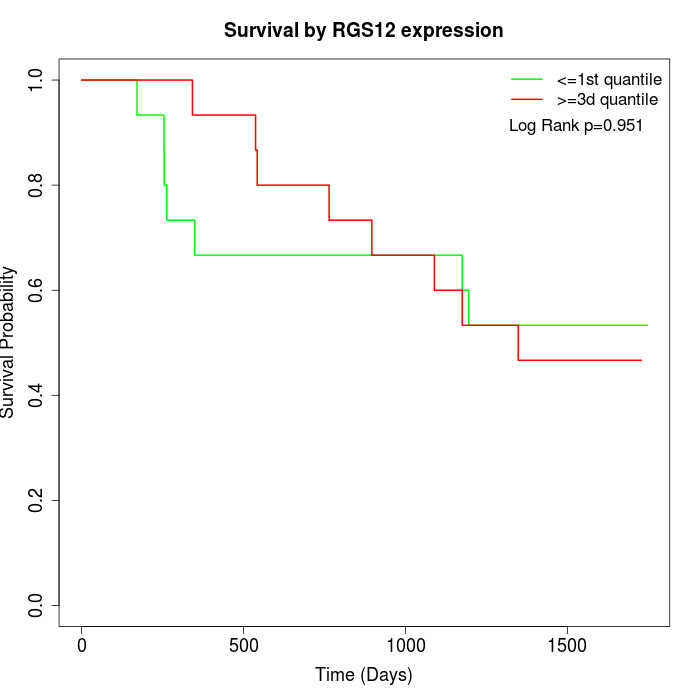
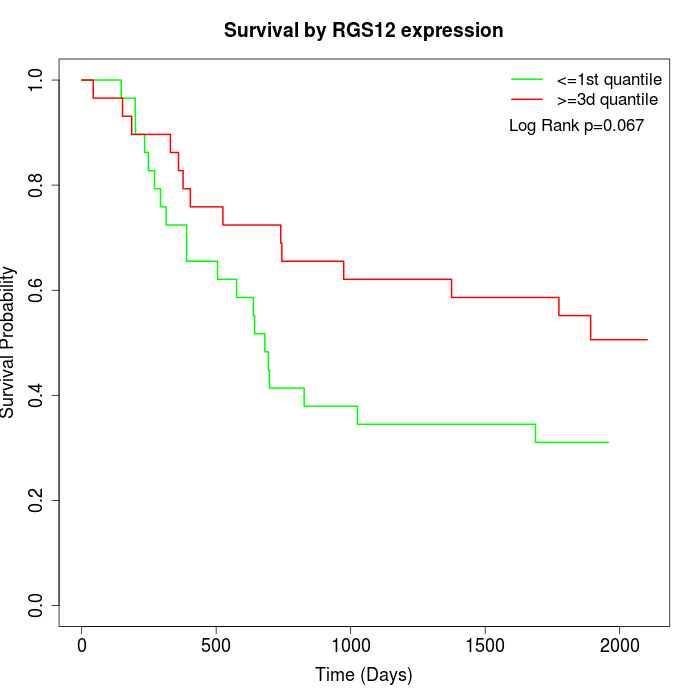
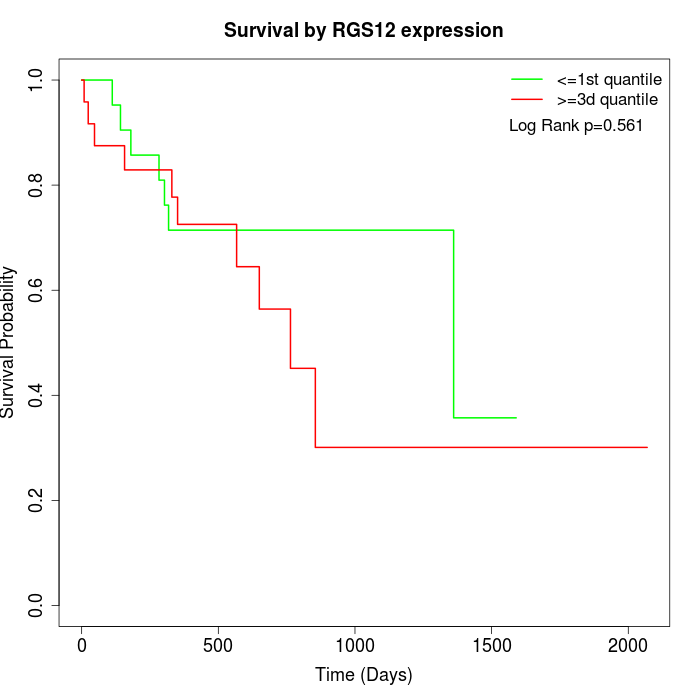
Survival by RGS12 expression:
Note: Click image to view full size file.
Copy number change of RGS12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS12 | 6002 | 0 | 17 | 13 | |
| GSE20123 | RGS12 | 6002 | 0 | 17 | 13 | |
| GSE43470 | RGS12 | 6002 | 0 | 18 | 25 | |
| GSE46452 | RGS12 | 6002 | 1 | 36 | 22 | |
| GSE47630 | RGS12 | 6002 | 1 | 19 | 20 | |
| GSE54993 | RGS12 | 6002 | 10 | 0 | 60 | |
| GSE54994 | RGS12 | 6002 | 3 | 12 | 38 | |
| GSE60625 | RGS12 | 6002 | 0 | 0 | 11 | |
| GSE74703 | RGS12 | 6002 | 0 | 14 | 22 | |
| GSE74704 | RGS12 | 6002 | 0 | 9 | 11 | |
| TCGA | RGS12 | 6002 | 7 | 47 | 42 |
Total number of gains: 22; Total number of losses: 189; Total Number of normals: 277.
Somatic mutations of RGS12:
Generating mutation plots.
Highly correlated genes for RGS12:
Showing top 20/645 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS12 | GRIPAP1 | 0.825554 | 3 | 0 | 3 |
| RGS12 | CDC42BPB | 0.811311 | 4 | 0 | 4 |
| RGS12 | MFSD2B | 0.771829 | 3 | 0 | 3 |
| RGS12 | TEN1 | 0.755739 | 3 | 0 | 3 |
| RGS12 | WDR55 | 0.745725 | 3 | 0 | 3 |
| RGS12 | FUT6 | 0.737495 | 6 | 0 | 6 |
| RGS12 | FAM163B | 0.725034 | 3 | 0 | 3 |
| RGS12 | KRTAP20-1 | 0.721385 | 3 | 0 | 3 |
| RGS12 | FANK1 | 0.71722 | 4 | 0 | 3 |
| RGS12 | RNF157 | 0.716969 | 3 | 0 | 3 |
| RGS12 | MAGIX | 0.716936 | 3 | 0 | 3 |
| RGS12 | OR2Z1 | 0.715135 | 3 | 0 | 3 |
| RGS12 | PTK6 | 0.71464 | 10 | 0 | 9 |
| RGS12 | OR10G8 | 0.711526 | 3 | 0 | 3 |
| RGS12 | CCDC157 | 0.71152 | 3 | 0 | 3 |
| RGS12 | NAT8B | 0.709664 | 3 | 0 | 3 |
| RGS12 | RIPK4 | 0.709006 | 8 | 0 | 7 |
| RGS12 | RARG | 0.708311 | 8 | 0 | 8 |
| RGS12 | PADI1 | 0.704155 | 6 | 0 | 6 |
| RGS12 | SEC13 | 0.703844 | 3 | 0 | 3 |
For details and further investigation, click here