| Full name: relaxin 2 | Alias Symbol: H2|RLXH2|bA12D24.1.1|bA12D24.1.2 | ||
| Type: protein-coding gene | Cytoband: 9p24.1 | ||
| Entrez ID: 6019 | HGNC ID: HGNC:10027 | Ensembl Gene: ENSG00000107014 | OMIM ID: 179740 |
| Drug and gene relationship at DGIdb | |||
Expression of RLN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RLN2 | 6019 | 214519_s_at | 0.3579 | 0.4199 | |
| GSE20347 | RLN2 | 6019 | 214519_s_at | -0.2359 | 0.0001 | |
| GSE23400 | RLN2 | 6019 | 214519_s_at | -0.0653 | 0.0141 | |
| GSE26886 | RLN2 | 6019 | 214519_s_at | 0.2500 | 0.4217 | |
| GSE29001 | RLN2 | 6019 | 214519_s_at | -0.1329 | 0.6974 | |
| GSE38129 | RLN2 | 6019 | 214519_s_at | -0.0376 | 0.7205 | |
| GSE45670 | RLN2 | 6019 | 214519_s_at | -0.0496 | 0.8076 | |
| GSE53622 | RLN2 | 6019 | 27061 | -0.7326 | 0.0000 | |
| GSE53624 | RLN2 | 6019 | 27061 | -0.5263 | 0.0137 | |
| GSE63941 | RLN2 | 6019 | 214519_s_at | 1.1162 | 0.2721 | |
| GSE77861 | RLN2 | 6019 | 214519_s_at | -0.1112 | 0.2769 | |
| TCGA | RLN2 | 6019 | RNAseq | 1.4980 | 0.0158 |
Upregulated datasets: 1; Downregulated datasets: 0.
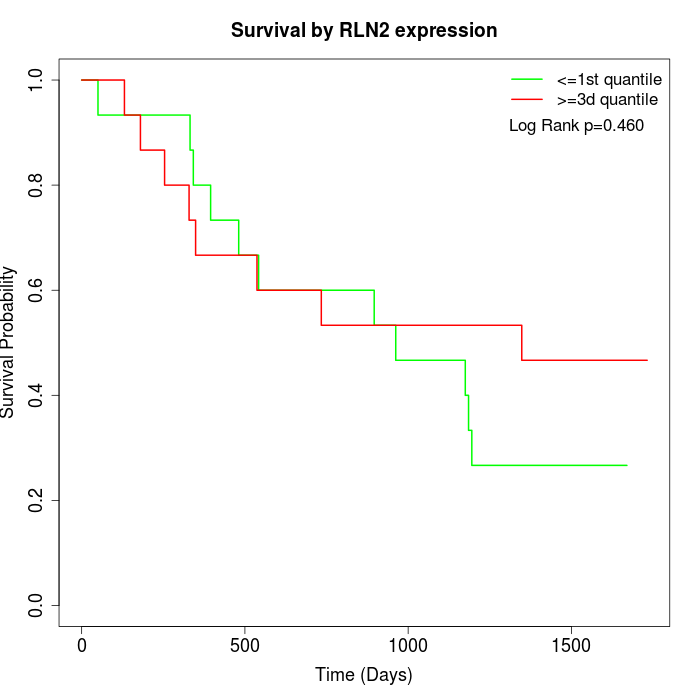
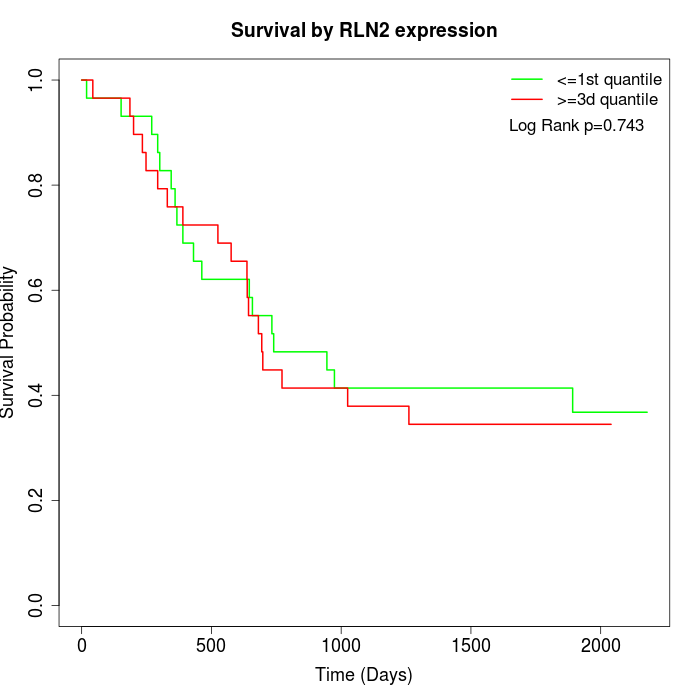
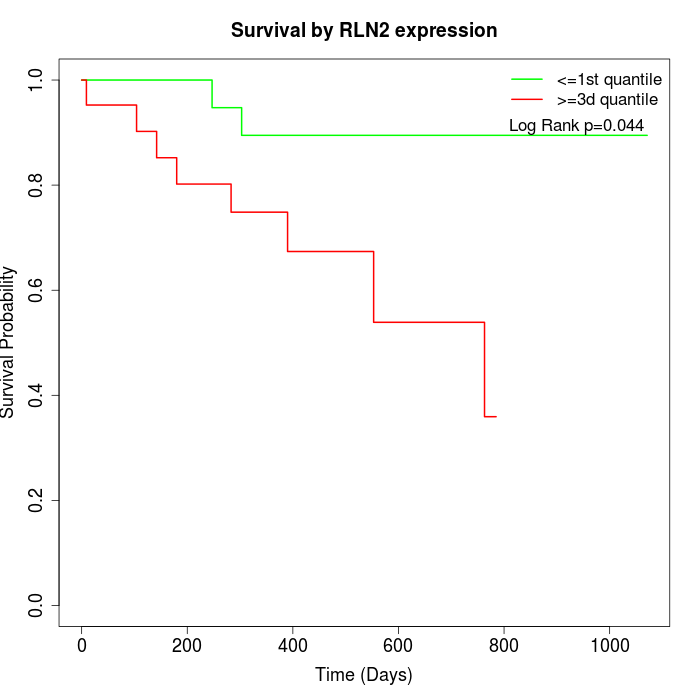
Survival by RLN2 expression:
Note: Click image to view full size file.
Copy number change of RLN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RLN2 | 6019 | 1 | 17 | 12 | |
| GSE20123 | RLN2 | 6019 | 1 | 16 | 13 | |
| GSE43470 | RLN2 | 6019 | 5 | 11 | 27 | |
| GSE46452 | RLN2 | 6019 | 2 | 23 | 34 | |
| GSE47630 | RLN2 | 6019 | 5 | 26 | 9 | |
| GSE54993 | RLN2 | 6019 | 6 | 3 | 61 | |
| GSE54994 | RLN2 | 6019 | 9 | 17 | 27 | |
| GSE60625 | RLN2 | 6019 | 0 | 2 | 9 | |
| GSE74703 | RLN2 | 6019 | 5 | 8 | 23 | |
| GSE74704 | RLN2 | 6019 | 0 | 12 | 8 | |
| TCGA | RLN2 | 6019 | 14 | 50 | 32 |
Total number of gains: 48; Total number of losses: 185; Total Number of normals: 255.
Somatic mutations of RLN2:
Generating mutation plots.
Highly correlated genes for RLN2:
Showing top 20/34 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RLN2 | IFT122 | 0.664313 | 3 | 0 | 3 |
| RLN2 | CPNE4 | 0.642819 | 3 | 0 | 3 |
| RLN2 | ZNF45 | 0.640468 | 4 | 0 | 3 |
| RLN2 | TCP11 | 0.63816 | 3 | 0 | 3 |
| RLN2 | ZNF627 | 0.625972 | 3 | 0 | 3 |
| RLN2 | SLAIN1 | 0.619036 | 5 | 0 | 4 |
| RLN2 | RLN1 | 0.611304 | 8 | 0 | 5 |
| RLN2 | ZNF234 | 0.606626 | 4 | 0 | 4 |
| RLN2 | ZNF180 | 0.586712 | 4 | 0 | 3 |
| RLN2 | CHRNB3 | 0.583752 | 3 | 0 | 3 |
| RLN2 | RERE | 0.581845 | 3 | 0 | 3 |
| RLN2 | TRIM8 | 0.578341 | 4 | 0 | 3 |
| RLN2 | ZNF112 | 0.578181 | 4 | 0 | 3 |
| RLN2 | LINC00240 | 0.575387 | 3 | 0 | 3 |
| RLN2 | PLCL2 | 0.57115 | 3 | 0 | 3 |
| RLN2 | CHAT | 0.570504 | 3 | 0 | 3 |
| RLN2 | ATP6V0E2 | 0.570165 | 4 | 0 | 3 |
| RLN2 | LRP1B | 0.569753 | 3 | 0 | 3 |
| RLN2 | FMO5 | 0.566372 | 3 | 0 | 3 |
| RLN2 | EFHC2 | 0.558879 | 3 | 0 | 3 |
For details and further investigation, click here