| Full name: arginine-glutamic acid dipeptide repeats | Alias Symbol: KIAA0458|ARP|ARG|DNB1 | ||
| Type: protein-coding gene | Cytoband: 1p36.23 | ||
| Entrez ID: 473 | HGNC ID: HGNC:9965 | Ensembl Gene: ENSG00000142599 | OMIM ID: 605226 |
| Drug and gene relationship at DGIdb | |||
Expression of RERE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RERE | 473 | 200940_s_at | -0.5375 | 0.5910 | |
| GSE20347 | RERE | 473 | 200940_s_at | -0.5284 | 0.0011 | |
| GSE23400 | RERE | 473 | 200940_s_at | -0.5682 | 0.0000 | |
| GSE26886 | RERE | 473 | 200940_s_at | -0.2228 | 0.5599 | |
| GSE29001 | RERE | 473 | 200940_s_at | -0.4944 | 0.0548 | |
| GSE38129 | RERE | 473 | 200940_s_at | -0.6647 | 0.0013 | |
| GSE45670 | RERE | 473 | 200940_s_at | -0.8815 | 0.0000 | |
| GSE53622 | RERE | 473 | 41665 | 0.0625 | 0.5028 | |
| GSE53624 | RERE | 473 | 41665 | 0.3826 | 0.0024 | |
| GSE63941 | RERE | 473 | 200940_s_at | -1.5207 | 0.0015 | |
| GSE77861 | RERE | 473 | 200940_s_at | -0.4610 | 0.1076 | |
| GSE97050 | RERE | A_21_P0011288 | -0.3159 | 0.4834 | ||
| SRP007169 | RERE | 473 | RNAseq | -0.5152 | 0.2074 | |
| SRP008496 | RERE | 473 | RNAseq | -0.6302 | 0.0271 | |
| SRP064894 | RERE | 473 | RNAseq | -0.5767 | 0.0004 | |
| SRP133303 | RERE | 473 | RNAseq | -0.6538 | 0.0008 | |
| SRP159526 | RERE | 473 | RNAseq | -0.7554 | 0.0092 | |
| SRP193095 | RERE | 473 | RNAseq | -0.2722 | 0.1174 | |
| SRP219564 | RERE | 473 | RNAseq | -0.3353 | 0.5583 | |
| TCGA | RERE | 473 | RNAseq | -0.3042 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
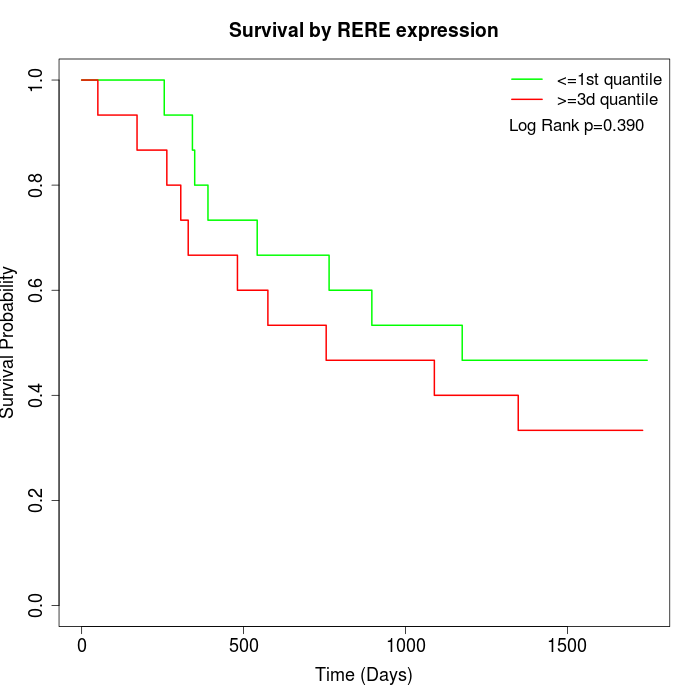
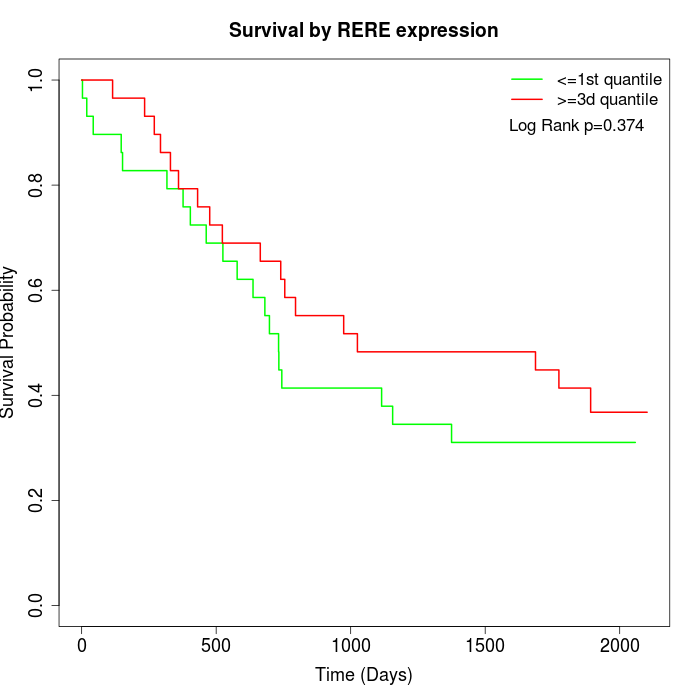
Survival by RERE expression:
Note: Click image to view full size file.
Copy number change of RERE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RERE | 473 | 1 | 5 | 24 | |
| GSE20123 | RERE | 473 | 1 | 3 | 26 | |
| GSE43470 | RERE | 473 | 6 | 5 | 32 | |
| GSE46452 | RERE | 473 | 7 | 1 | 51 | |
| GSE47630 | RERE | 473 | 8 | 4 | 28 | |
| GSE54993 | RERE | 473 | 3 | 2 | 65 | |
| GSE54994 | RERE | 473 | 13 | 3 | 37 | |
| GSE60625 | RERE | 473 | 0 | 0 | 11 | |
| GSE74703 | RERE | 473 | 5 | 3 | 28 | |
| GSE74704 | RERE | 473 | 1 | 0 | 19 | |
| TCGA | RERE | 473 | 12 | 24 | 60 |
Total number of gains: 57; Total number of losses: 50; Total Number of normals: 381.
Somatic mutations of RERE:
Generating mutation plots.
Highly correlated genes for RERE:
Showing top 20/1108 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RERE | RBPMS2 | 0.797071 | 3 | 0 | 3 |
| RERE | EPHA3 | 0.793336 | 4 | 0 | 4 |
| RERE | CDC16 | 0.790243 | 3 | 0 | 3 |
| RERE | FILIP1 | 0.783773 | 3 | 0 | 3 |
| RERE | FRMD3 | 0.769274 | 3 | 0 | 3 |
| RERE | C1QTNF7 | 0.766247 | 3 | 0 | 3 |
| RERE | ANGPTL1 | 0.760376 | 4 | 0 | 4 |
| RERE | PIN1 | 0.759041 | 3 | 0 | 3 |
| RERE | TMEM220 | 0.751513 | 3 | 0 | 3 |
| RERE | MAOB | 0.749344 | 7 | 0 | 7 |
| RERE | KIAA1191 | 0.744794 | 4 | 0 | 4 |
| RERE | MAMDC2 | 0.743753 | 5 | 0 | 5 |
| RERE | PRELP | 0.743421 | 5 | 0 | 5 |
| RERE | RAB9B | 0.74193 | 4 | 0 | 4 |
| RERE | JAM3 | 0.739068 | 6 | 0 | 6 |
| RERE | INMT | 0.736236 | 4 | 0 | 4 |
| RERE | ATXN7L3B | 0.73319 | 3 | 0 | 3 |
| RERE | TMEM178A | 0.732387 | 3 | 0 | 3 |
| RERE | RRAGB | 0.731669 | 4 | 0 | 4 |
| RERE | GFRA1 | 0.72503 | 4 | 0 | 4 |
For details and further investigation, click here