| Full name: relaxin 1 | Alias Symbol: H1 | ||
| Type: protein-coding gene | Cytoband: 9p24.1 | ||
| Entrez ID: 6013 | HGNC ID: HGNC:10026 | Ensembl Gene: ENSG00000107018 | OMIM ID: 179730 |
| Drug and gene relationship at DGIdb | |||
Expression of RLN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RLN1 | 6013 | 211753_s_at | -0.0714 | 0.8454 | |
| GSE20347 | RLN1 | 6013 | 211753_s_at | -0.0604 | 0.3656 | |
| GSE23400 | RLN1 | 6013 | 211753_s_at | -0.1759 | 0.0000 | |
| GSE26886 | RLN1 | 6013 | 211753_s_at | 0.1294 | 0.3751 | |
| GSE29001 | RLN1 | 6013 | 211753_s_at | -0.0038 | 0.9808 | |
| GSE38129 | RLN1 | 6013 | 211753_s_at | -0.0733 | 0.2748 | |
| GSE45670 | RLN1 | 6013 | 211753_s_at | -0.0622 | 0.4811 | |
| GSE53622 | RLN1 | 6013 | 114855 | -0.4186 | 0.0024 | |
| GSE53624 | RLN1 | 6013 | 114855 | -0.2600 | 0.1376 | |
| GSE63941 | RLN1 | 6013 | 211753_s_at | 0.1597 | 0.5495 | |
| GSE77861 | RLN1 | 6013 | 211753_s_at | 0.0016 | 0.9845 | |
| TCGA | RLN1 | 6013 | RNAseq | -0.0976 | 0.8886 |
Upregulated datasets: 0; Downregulated datasets: 0.
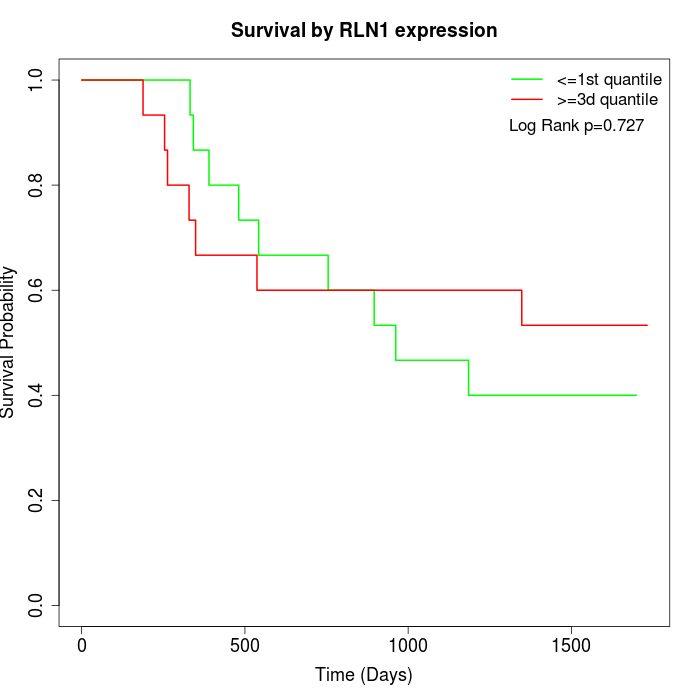
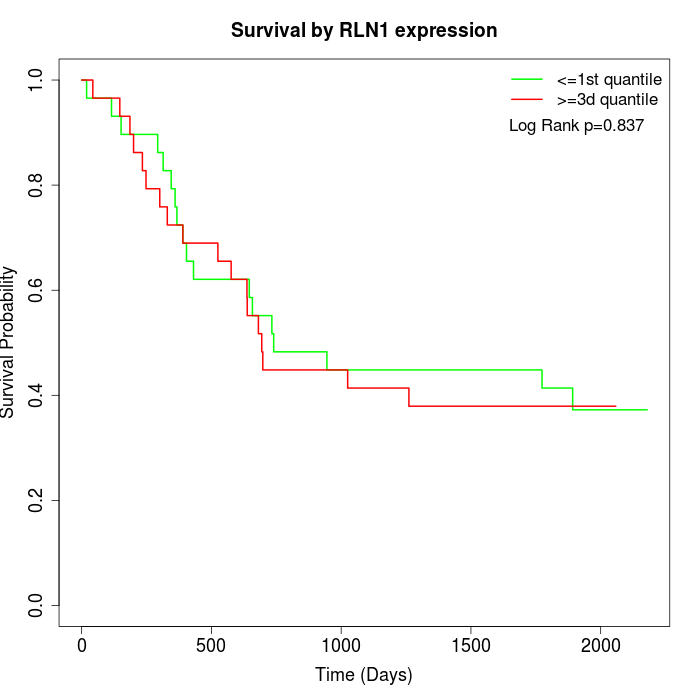
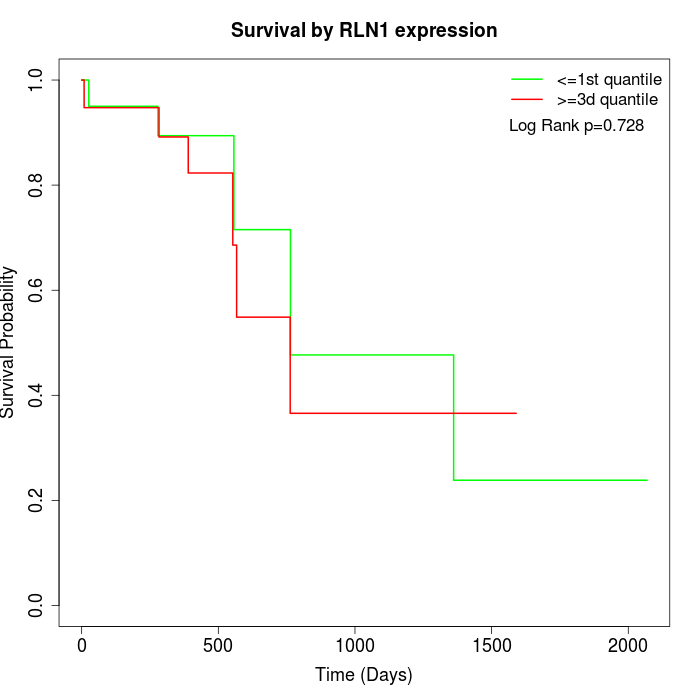
Survival by RLN1 expression:
Note: Click image to view full size file.
Copy number change of RLN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RLN1 | 6013 | 1 | 17 | 12 | |
| GSE20123 | RLN1 | 6013 | 1 | 17 | 12 | |
| GSE43470 | RLN1 | 6013 | 5 | 11 | 27 | |
| GSE46452 | RLN1 | 6013 | 2 | 23 | 34 | |
| GSE47630 | RLN1 | 6013 | 5 | 26 | 9 | |
| GSE54993 | RLN1 | 6013 | 6 | 3 | 61 | |
| GSE54994 | RLN1 | 6013 | 9 | 17 | 27 | |
| GSE60625 | RLN1 | 6013 | 0 | 2 | 9 | |
| GSE74703 | RLN1 | 6013 | 5 | 8 | 23 | |
| GSE74704 | RLN1 | 6013 | 0 | 13 | 7 | |
| TCGA | RLN1 | 6013 | 14 | 50 | 32 |
Total number of gains: 48; Total number of losses: 187; Total Number of normals: 253.
Somatic mutations of RLN1:
Generating mutation plots.
Highly correlated genes for RLN1:
Showing top 20/27 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RLN1 | WFDC2 | 0.707466 | 3 | 0 | 3 |
| RLN1 | CD244 | 0.612754 | 3 | 0 | 3 |
| RLN1 | RLN2 | 0.611304 | 8 | 0 | 5 |
| RLN1 | RUNX1T1 | 0.59986 | 3 | 0 | 3 |
| RLN1 | MYEF2 | 0.598164 | 3 | 0 | 3 |
| RLN1 | LGALS13 | 0.597212 | 3 | 0 | 3 |
| RLN1 | TMEM71 | 0.586776 | 4 | 0 | 3 |
| RLN1 | ENPP5 | 0.582739 | 4 | 0 | 3 |
| RLN1 | EYA1 | 0.580076 | 4 | 0 | 3 |
| RLN1 | NT5M | 0.568792 | 3 | 0 | 3 |
| RLN1 | BTN1A1 | 0.56502 | 3 | 0 | 3 |
| RLN1 | LRRC31 | 0.553919 | 4 | 0 | 3 |
| RLN1 | C8orf34 | 0.549171 | 4 | 0 | 4 |
| RLN1 | PYHIN1 | 0.548278 | 3 | 0 | 3 |
| RLN1 | RBFADN | 0.548205 | 4 | 0 | 3 |
| RLN1 | LIN7A | 0.544862 | 6 | 0 | 4 |
| RLN1 | SMAD9 | 0.536422 | 4 | 0 | 3 |
| RLN1 | LCT | 0.528897 | 6 | 0 | 3 |
| RLN1 | CYFIP2 | 0.528697 | 6 | 0 | 3 |
| RLN1 | TRIM52-AS1 | 0.528012 | 5 | 0 | 3 |
For details and further investigation, click here