| Full name: zinc finger protein 45 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 7596 | HGNC ID: HGNC:13111 | Ensembl Gene: ENSG00000124459 | OMIM ID: 194554 |
| Drug and gene relationship at DGIdb | |||
Expression of ZNF45:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZNF45 | 7596 | 222028_at | -0.0264 | 0.9714 | |
| GSE20347 | ZNF45 | 7596 | 222028_at | 0.0941 | 0.6186 | |
| GSE23400 | ZNF45 | 7596 | 222028_at | 0.1850 | 0.0322 | |
| GSE26886 | ZNF45 | 7596 | 222028_at | 0.5826 | 0.0971 | |
| GSE29001 | ZNF45 | 7596 | 222028_at | 0.4561 | 0.3529 | |
| GSE38129 | ZNF45 | 7596 | 222028_at | 0.0492 | 0.7453 | |
| GSE45670 | ZNF45 | 7596 | 222028_at | -0.1288 | 0.4613 | |
| GSE53622 | ZNF45 | 7596 | 37825 | -0.0702 | 0.4367 | |
| GSE53624 | ZNF45 | 7596 | 37825 | 0.0034 | 0.9723 | |
| GSE63941 | ZNF45 | 7596 | 222028_at | -0.8452 | 0.0748 | |
| GSE77861 | ZNF45 | 7596 | 222028_at | 0.2249 | 0.4199 | |
| GSE97050 | ZNF45 | 7596 | A_23_P426472 | -0.1168 | 0.7715 | |
| SRP007169 | ZNF45 | 7596 | RNAseq | 0.1858 | 0.6317 | |
| SRP008496 | ZNF45 | 7596 | RNAseq | 0.2614 | 0.3606 | |
| SRP064894 | ZNF45 | 7596 | RNAseq | 0.0951 | 0.6356 | |
| SRP133303 | ZNF45 | 7596 | RNAseq | 0.1271 | 0.4183 | |
| SRP159526 | ZNF45 | 7596 | RNAseq | 0.2661 | 0.3516 | |
| SRP193095 | ZNF45 | 7596 | RNAseq | -0.0144 | 0.8892 | |
| SRP219564 | ZNF45 | 7596 | RNAseq | -0.2986 | 0.4269 | |
| TCGA | ZNF45 | 7596 | RNAseq | -0.1476 | 0.0303 |
Upregulated datasets: 0; Downregulated datasets: 0.
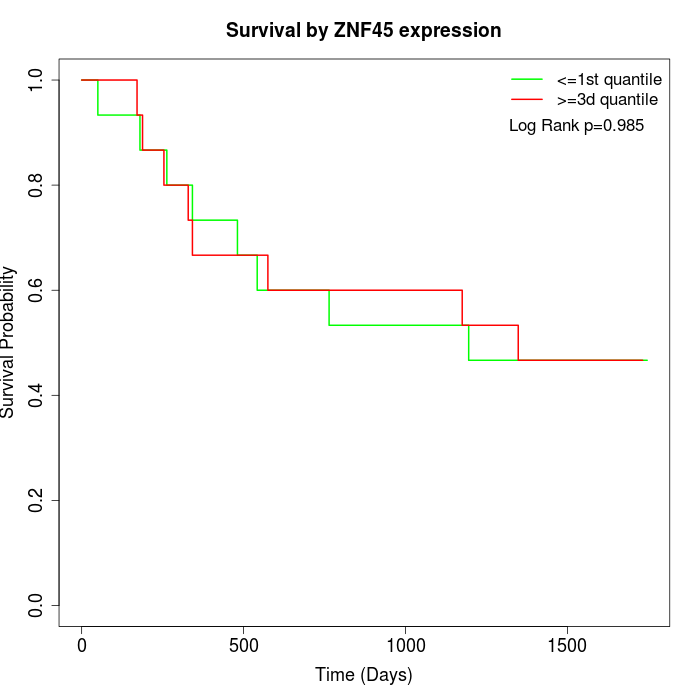
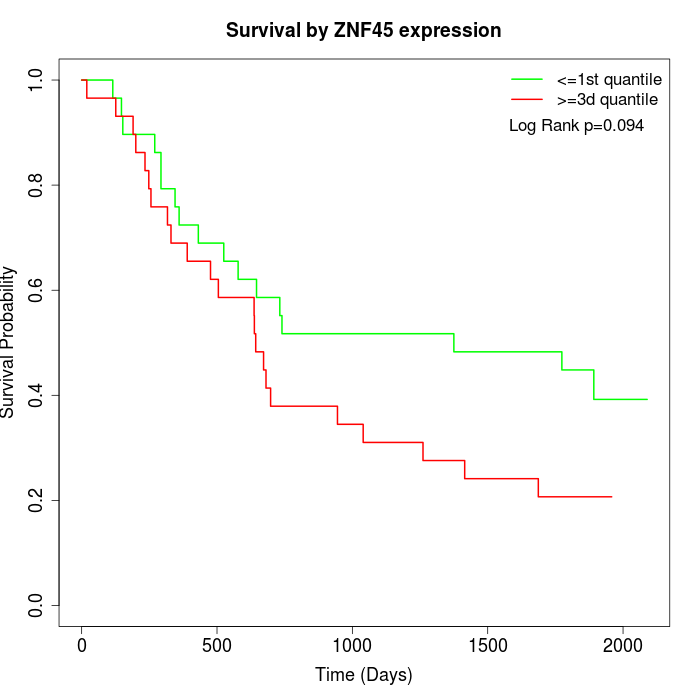
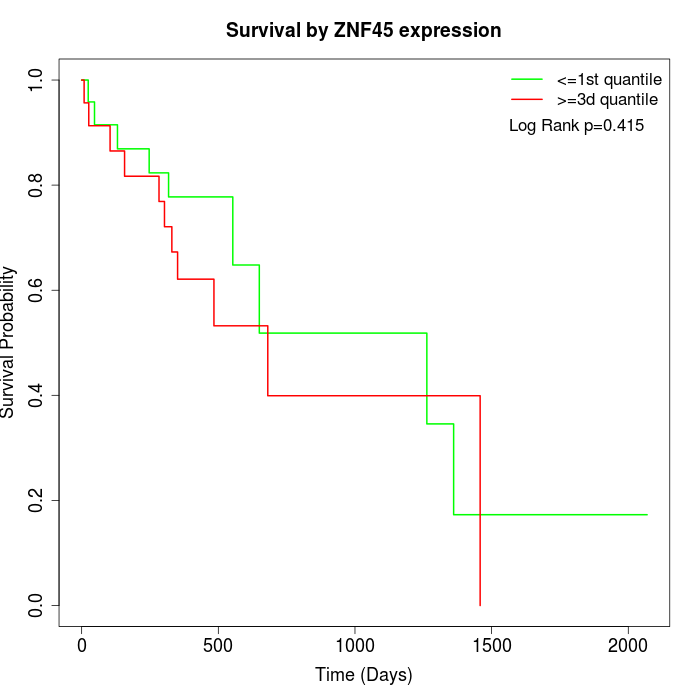
Survival by ZNF45 expression:
Note: Click image to view full size file.
Copy number change of ZNF45:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZNF45 | 7596 | 6 | 5 | 19 | |
| GSE20123 | ZNF45 | 7596 | 5 | 4 | 21 | |
| GSE43470 | ZNF45 | 7596 | 4 | 11 | 28 | |
| GSE46452 | ZNF45 | 7596 | 45 | 1 | 13 | |
| GSE47630 | ZNF45 | 7596 | 8 | 6 | 26 | |
| GSE54993 | ZNF45 | 7596 | 17 | 4 | 49 | |
| GSE54994 | ZNF45 | 7596 | 6 | 12 | 35 | |
| GSE60625 | ZNF45 | 7596 | 9 | 0 | 2 | |
| GSE74703 | ZNF45 | 7596 | 4 | 7 | 25 | |
| GSE74704 | ZNF45 | 7596 | 4 | 2 | 14 | |
| TCGA | ZNF45 | 7596 | 14 | 16 | 66 |
Total number of gains: 122; Total number of losses: 68; Total Number of normals: 298.
Somatic mutations of ZNF45:
Generating mutation plots.
Highly correlated genes for ZNF45:
Showing top 20/325 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZNF45 | ZFP3 | 0.693709 | 3 | 0 | 3 |
| ZNF45 | ELF2 | 0.682181 | 4 | 0 | 3 |
| ZNF45 | DDX42 | 0.681011 | 3 | 0 | 3 |
| ZNF45 | ZNF805 | 0.667009 | 7 | 0 | 5 |
| ZNF45 | ZNF780B | 0.665138 | 4 | 0 | 3 |
| ZNF45 | UTP23 | 0.658983 | 3 | 0 | 3 |
| ZNF45 | CHIC1 | 0.658134 | 4 | 0 | 3 |
| ZNF45 | BDNF | 0.656654 | 3 | 0 | 3 |
| ZNF45 | TOPORS | 0.656175 | 4 | 0 | 3 |
| ZNF45 | PAIP1 | 0.655167 | 5 | 0 | 5 |
| ZNF45 | ZFYVE1 | 0.653872 | 3 | 0 | 3 |
| ZNF45 | DPY19L2 | 0.653789 | 3 | 0 | 3 |
| ZNF45 | SHPRH | 0.649793 | 4 | 0 | 3 |
| ZNF45 | PNKD | 0.646909 | 3 | 0 | 3 |
| ZNF45 | MDM4 | 0.644921 | 4 | 0 | 3 |
| ZNF45 | NCBP1 | 0.641394 | 3 | 0 | 3 |
| ZNF45 | ATG14 | 0.640484 | 4 | 0 | 4 |
| ZNF45 | RLN2 | 0.640468 | 4 | 0 | 3 |
| ZNF45 | ZNF112 | 0.635039 | 11 | 0 | 10 |
| ZNF45 | AGPS | 0.634794 | 3 | 0 | 3 |
For details and further investigation, click here