| Full name: S-antigen visual arrestin | Alias Symbol: ARRESTIN|RP47 | ||
| Type: protein-coding gene | Cytoband: 2q37.1 | ||
| Entrez ID: 6295 | HGNC ID: HGNC:10521 | Ensembl Gene: ENSG00000130561 | OMIM ID: 181031 |
| Drug and gene relationship at DGIdb | |||
Expression of SAG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SAG | 6295 | 206671_at | 0.1245 | 0.7346 | |
| GSE20347 | SAG | 6295 | 206671_at | 0.0515 | 0.6157 | |
| GSE23400 | SAG | 6295 | 206671_at | -0.0316 | 0.4883 | |
| GSE26886 | SAG | 6295 | 206671_at | 0.1431 | 0.2509 | |
| GSE29001 | SAG | 6295 | 206671_at | -0.0043 | 0.9840 | |
| GSE38129 | SAG | 6295 | 206671_at | -0.0548 | 0.6362 | |
| GSE45670 | SAG | 6295 | 206671_at | 0.1413 | 0.0800 | |
| GSE53622 | SAG | 6295 | 61640 | 0.2416 | 0.1119 | |
| GSE53624 | SAG | 6295 | 61640 | 0.1661 | 0.3487 | |
| GSE63941 | SAG | 6295 | 206671_at | 0.1511 | 0.4065 | |
| GSE77861 | SAG | 6295 | 206671_at | 0.1015 | 0.3115 | |
| GSE97050 | SAG | 6295 | A_33_P3232203 | -0.4275 | 0.2764 |
Upregulated datasets: 0; Downregulated datasets: 0.
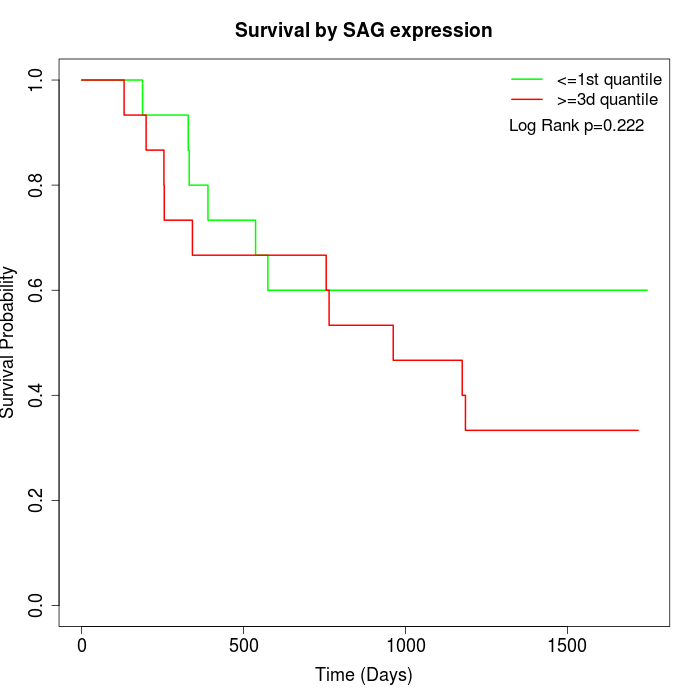
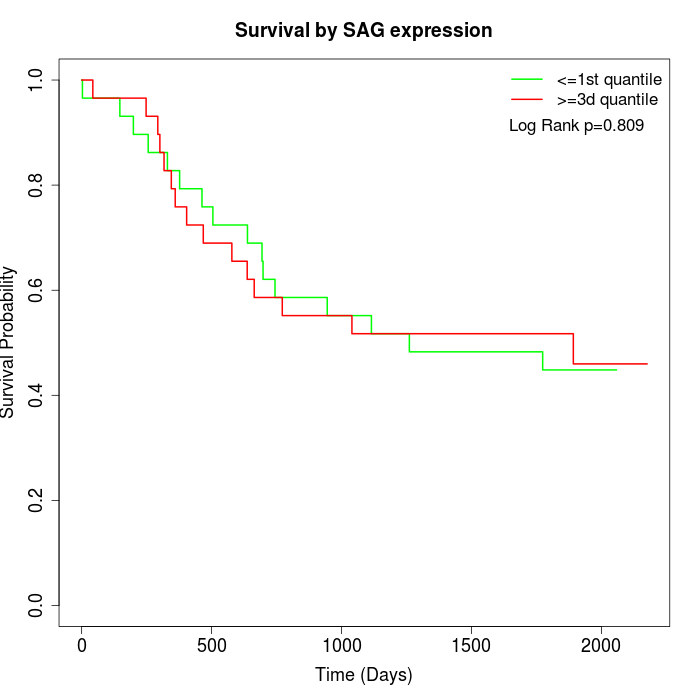
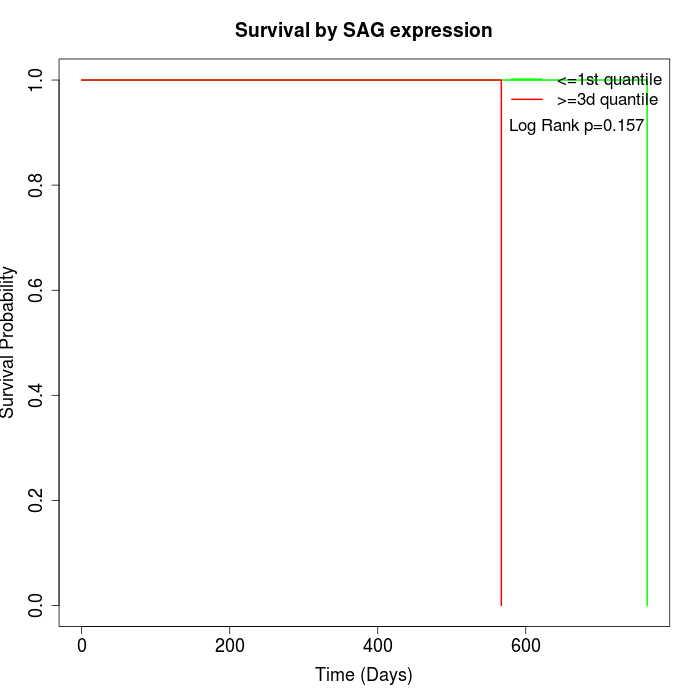
Survival by SAG expression:
Note: Click image to view full size file.
Copy number change of SAG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SAG | 6295 | 1 | 12 | 17 | |
| GSE20123 | SAG | 6295 | 1 | 12 | 17 | |
| GSE43470 | SAG | 6295 | 1 | 9 | 33 | |
| GSE46452 | SAG | 6295 | 0 | 5 | 54 | |
| GSE47630 | SAG | 6295 | 4 | 5 | 31 | |
| GSE54993 | SAG | 6295 | 3 | 2 | 65 | |
| GSE54994 | SAG | 6295 | 6 | 10 | 37 | |
| GSE60625 | SAG | 6295 | 0 | 3 | 8 | |
| GSE74703 | SAG | 6295 | 1 | 7 | 28 | |
| GSE74704 | SAG | 6295 | 1 | 5 | 14 | |
| TCGA | SAG | 6295 | 12 | 28 | 56 |
Total number of gains: 30; Total number of losses: 98; Total Number of normals: 360.
Somatic mutations of SAG:
Generating mutation plots.
Highly correlated genes for SAG:
Showing top 20/615 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SAG | PEX14 | 0.82224 | 4 | 0 | 4 |
| SAG | ADAM22 | 0.791331 | 5 | 0 | 5 |
| SAG | EEF1D | 0.764796 | 3 | 0 | 3 |
| SAG | CDON | 0.763653 | 5 | 0 | 5 |
| SAG | NELL1 | 0.759618 | 3 | 0 | 3 |
| SAG | SP100 | 0.7568 | 3 | 0 | 3 |
| SAG | GAL | 0.751809 | 3 | 0 | 3 |
| SAG | NTNG1 | 0.749164 | 4 | 0 | 4 |
| SAG | G6PC2 | 0.74608 | 4 | 0 | 4 |
| SAG | CHML | 0.73463 | 3 | 0 | 3 |
| SAG | PRPS1L1 | 0.72686 | 6 | 0 | 6 |
| SAG | ZC3H14 | 0.7209 | 3 | 0 | 3 |
| SAG | GABRR2 | 0.714813 | 6 | 0 | 5 |
| SAG | DNAJA4 | 0.711852 | 4 | 0 | 4 |
| SAG | UHRF1BP1 | 0.709977 | 3 | 0 | 3 |
| SAG | TNPO1 | 0.708039 | 5 | 0 | 5 |
| SAG | SST | 0.705347 | 3 | 0 | 3 |
| SAG | RIMKLB | 0.703983 | 3 | 0 | 3 |
| SAG | P2RX6 | 0.703453 | 4 | 0 | 4 |
| SAG | FANCC | 0.703057 | 4 | 0 | 4 |
For details and further investigation, click here