| Full name: SHANK2 antisense RNA 3 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 11q13.4 | ||
| Entrez ID: 220070 | HGNC ID: HGNC:25098 | Ensembl Gene: ENSG00000171671 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SHANK2-AS3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SHANK2-AS3 | 220070 | 224139_at | 0.0456 | 0.8651 | |
| GSE26886 | SHANK2-AS3 | 220070 | 224139_at | -0.0143 | 0.8867 | |
| GSE45670 | SHANK2-AS3 | 220070 | 224139_at | 0.0509 | 0.4849 | |
| GSE53622 | SHANK2-AS3 | 220070 | 547 | 0.1031 | 0.2215 | |
| GSE53624 | SHANK2-AS3 | 220070 | 547 | 0.0661 | 0.5265 | |
| GSE63941 | SHANK2-AS3 | 220070 | 224139_at | 0.1465 | 0.2195 | |
| GSE77861 | SHANK2-AS3 | 220070 | 224139_at | -0.0165 | 0.9006 | |
| SRP133303 | SHANK2-AS3 | 220070 | RNAseq | 0.5993 | 0.0010 | |
| SRP159526 | SHANK2-AS3 | 220070 | RNAseq | 0.8564 | 0.1067 | |
| SRP193095 | SHANK2-AS3 | 220070 | RNAseq | 0.4159 | 0.0213 | |
| SRP219564 | SHANK2-AS3 | 220070 | RNAseq | 0.1640 | 0.6812 |
Upregulated datasets: 0; Downregulated datasets: 0.
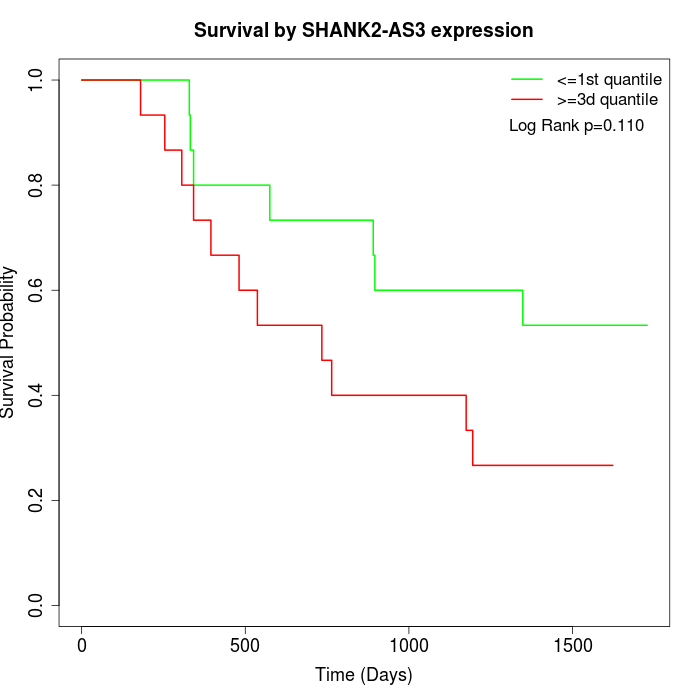
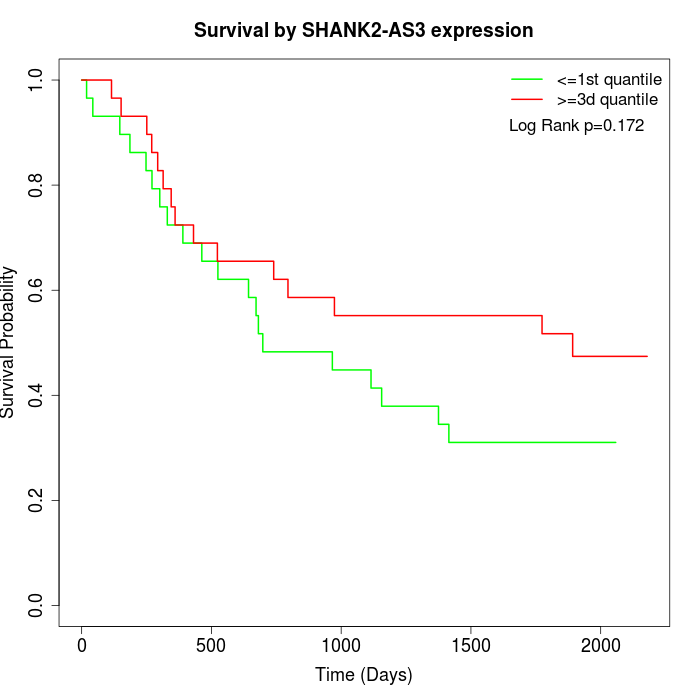
Survival by SHANK2-AS3 expression:
Note: Click image to view full size file.
Copy number change of SHANK2-AS3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SHANK2-AS3 | 220070 | 14 | 4 | 12 | |
| GSE20123 | SHANK2-AS3 | 220070 | 14 | 4 | 12 | |
| GSE43470 | SHANK2-AS3 | 220070 | 9 | 4 | 30 | |
| GSE46452 | SHANK2-AS3 | 220070 | 24 | 7 | 28 | |
| GSE47630 | SHANK2-AS3 | 220070 | 14 | 7 | 19 | |
| GSE54993 | SHANK2-AS3 | 220070 | 2 | 17 | 51 | |
| GSE54994 | SHANK2-AS3 | 220070 | 18 | 10 | 25 | |
| GSE60625 | SHANK2-AS3 | 220070 | 0 | 8 | 3 | |
| GSE74703 | SHANK2-AS3 | 220070 | 7 | 3 | 26 | |
| GSE74704 | SHANK2-AS3 | 220070 | 10 | 2 | 8 |
Total number of gains: 112; Total number of losses: 66; Total Number of normals: 214.
Somatic mutations of SHANK2-AS3:
Generating mutation plots.
Highly correlated genes for SHANK2-AS3:
Showing top 20/84 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SHANK2-AS3 | LINC00598 | 0.743986 | 3 | 0 | 3 |
| SHANK2-AS3 | LINC00475 | 0.715236 | 3 | 0 | 3 |
| SHANK2-AS3 | KCP | 0.713519 | 3 | 0 | 3 |
| SHANK2-AS3 | AGXT | 0.699333 | 3 | 0 | 3 |
| SHANK2-AS3 | CYP11B2 | 0.696036 | 3 | 0 | 3 |
| SHANK2-AS3 | CREB3L3 | 0.677756 | 3 | 0 | 3 |
| SHANK2-AS3 | SLC25A53 | 0.666163 | 3 | 0 | 3 |
| SHANK2-AS3 | SOX3 | 0.663397 | 3 | 0 | 3 |
| SHANK2-AS3 | STMN4 | 0.661323 | 4 | 0 | 4 |
| SHANK2-AS3 | HMX1 | 0.660861 | 4 | 0 | 4 |
| SHANK2-AS3 | EDA | 0.659967 | 3 | 0 | 3 |
| SHANK2-AS3 | KCNH3 | 0.654208 | 3 | 0 | 3 |
| SHANK2-AS3 | RIMBP2 | 0.65033 | 3 | 0 | 3 |
| SHANK2-AS3 | KLF16 | 0.64656 | 5 | 0 | 4 |
| SHANK2-AS3 | SEMA6C | 0.644595 | 4 | 0 | 3 |
| SHANK2-AS3 | KCTD19 | 0.637004 | 3 | 0 | 3 |
| SHANK2-AS3 | TNNI2 | 0.634454 | 3 | 0 | 3 |
| SHANK2-AS3 | NYX | 0.630364 | 4 | 0 | 4 |
| SHANK2-AS3 | UTF1 | 0.626067 | 4 | 0 | 4 |
| SHANK2-AS3 | RSPH9 | 0.624726 | 3 | 0 | 3 |
For details and further investigation, click here