| Full name: potassium channel tetramerization domain containing 19 | Alias Symbol: FLJ40162 | ||
| Type: protein-coding gene | Cytoband: 16q22.1 | ||
| Entrez ID: 146212 | HGNC ID: HGNC:24753 | Ensembl Gene: ENSG00000168676 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD19:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD19 | 146212 | 244538_at | -0.0738 | 0.7433 | |
| GSE26886 | KCTD19 | 146212 | 244538_at | 0.1175 | 0.2747 | |
| GSE45670 | KCTD19 | 146212 | 244538_at | 0.0849 | 0.4271 | |
| GSE53622 | KCTD19 | 146212 | 1927 | 0.1091 | 0.2249 | |
| GSE53624 | KCTD19 | 146212 | 1927 | -0.0924 | 0.4097 | |
| GSE63941 | KCTD19 | 146212 | 244538_at | -0.0586 | 0.5553 | |
| GSE77861 | KCTD19 | 146212 | 244538_at | 0.0029 | 0.9831 | |
| GSE97050 | KCTD19 | 146212 | A_33_P3255131 | 0.1455 | 0.6268 | |
| TCGA | KCTD19 | 146212 | RNAseq | 0.7732 | 0.2348 |
Upregulated datasets: 0; Downregulated datasets: 0.
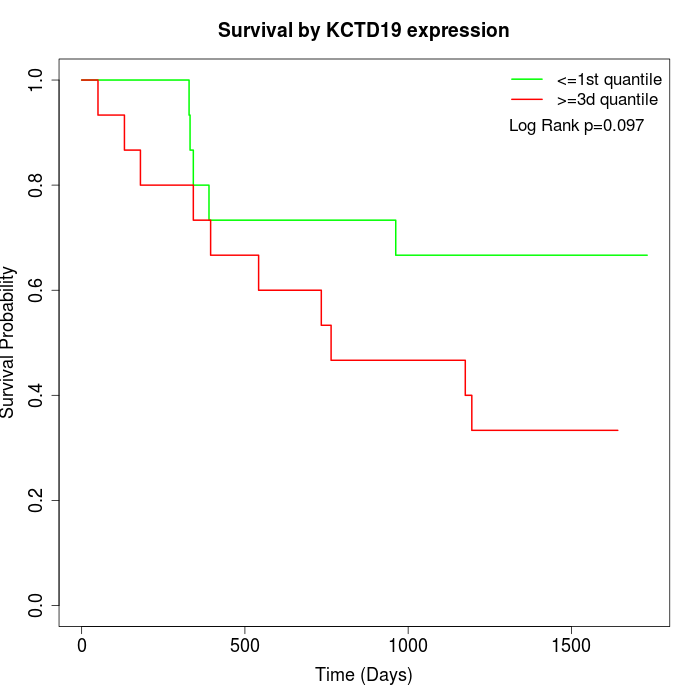
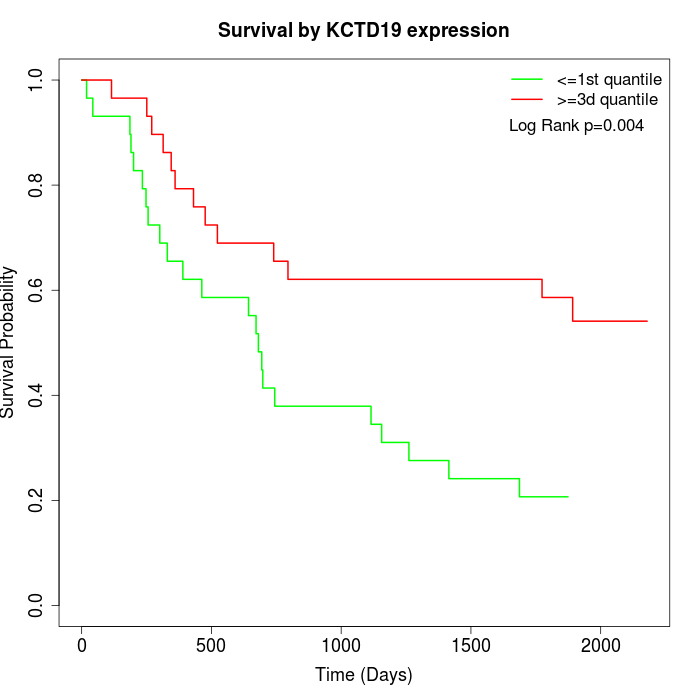
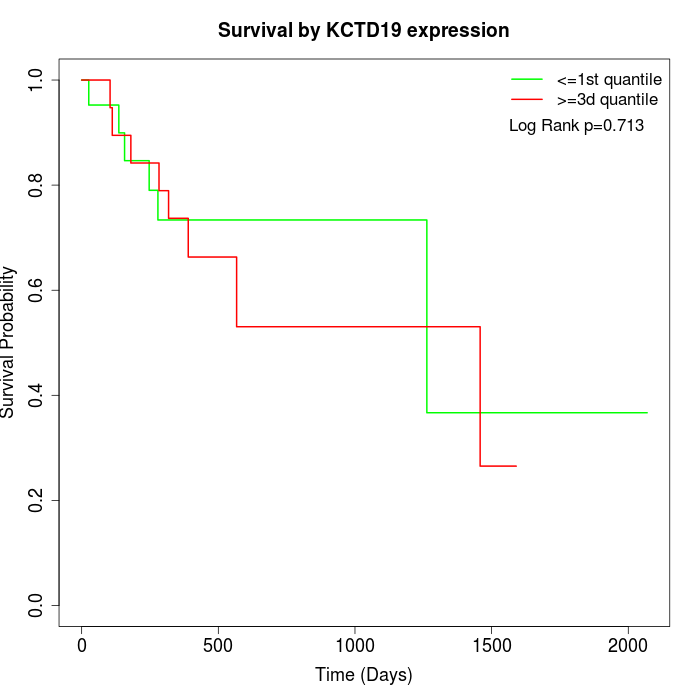
Survival by KCTD19 expression:
Note: Click image to view full size file.
Copy number change of KCTD19:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD19 | 146212 | 9 | 2 | 19 | |
| GSE20123 | KCTD19 | 146212 | 9 | 2 | 19 | |
| GSE43470 | KCTD19 | 146212 | 1 | 9 | 33 | |
| GSE46452 | KCTD19 | 146212 | 38 | 1 | 20 | |
| GSE47630 | KCTD19 | 146212 | 10 | 8 | 22 | |
| GSE54993 | KCTD19 | 146212 | 2 | 4 | 64 | |
| GSE54994 | KCTD19 | 146212 | 8 | 10 | 35 | |
| GSE60625 | KCTD19 | 146212 | 4 | 0 | 7 | |
| GSE74703 | KCTD19 | 146212 | 1 | 6 | 29 | |
| GSE74704 | KCTD19 | 146212 | 5 | 1 | 14 | |
| TCGA | KCTD19 | 146212 | 29 | 12 | 55 |
Total number of gains: 116; Total number of losses: 55; Total Number of normals: 317.
Somatic mutations of KCTD19:
Generating mutation plots.
Highly correlated genes for KCTD19:
Showing top 20/363 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD19 | SPACA3 | 0.848658 | 3 | 0 | 3 |
| KCTD19 | PANX2 | 0.82076 | 3 | 0 | 3 |
| KCTD19 | GRIK4 | 0.820373 | 3 | 0 | 3 |
| KCTD19 | CD72 | 0.800307 | 3 | 0 | 3 |
| KCTD19 | REN | 0.796363 | 3 | 0 | 3 |
| KCTD19 | STRC | 0.792813 | 3 | 0 | 3 |
| KCTD19 | BEST3 | 0.774642 | 4 | 0 | 4 |
| KCTD19 | MYOZ3 | 0.771297 | 3 | 0 | 3 |
| KCTD19 | OR4D6 | 0.770202 | 3 | 0 | 3 |
| KCTD19 | ACE | 0.766185 | 3 | 0 | 3 |
| KCTD19 | FGF22 | 0.755874 | 3 | 0 | 3 |
| KCTD19 | LHX9 | 0.753843 | 3 | 0 | 3 |
| KCTD19 | C9orf153 | 0.752968 | 3 | 0 | 3 |
| KCTD19 | GPR152 | 0.749831 | 3 | 0 | 3 |
| KCTD19 | MC3R | 0.7358 | 4 | 0 | 4 |
| KCTD19 | ALX4 | 0.735785 | 4 | 0 | 4 |
| KCTD19 | TMEM235 | 0.734466 | 3 | 0 | 3 |
| KCTD19 | FZD9 | 0.731741 | 3 | 0 | 3 |
| KCTD19 | BSX | 0.729968 | 3 | 0 | 3 |
| KCTD19 | TNFRSF14 | 0.726247 | 3 | 0 | 3 |
For details and further investigation, click here