| Full name: SLAM family member 9 | Alias Symbol: CD2F-10|CD84-H1|SF2001 | ||
| Type: protein-coding gene | Cytoband: 1q23.2 | ||
| Entrez ID: 89886 | HGNC ID: HGNC:18430 | Ensembl Gene: ENSG00000162723 | OMIM ID: 608589 |
| Drug and gene relationship at DGIdb | |||
Expression of SLAMF9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SLAMF9 | 89886 | 1553770_a_at | 0.2413 | 0.5303 | |
| GSE26886 | SLAMF9 | 89886 | 1553770_a_at | 0.2688 | 0.1586 | |
| GSE45670 | SLAMF9 | 89886 | 1553770_a_at | 0.6783 | 0.0442 | |
| GSE53622 | SLAMF9 | 89886 | 101174 | 0.2186 | 0.0188 | |
| GSE53624 | SLAMF9 | 89886 | 101174 | 0.2749 | 0.0058 | |
| GSE63941 | SLAMF9 | 89886 | 1553770_a_at | 0.1931 | 0.1249 | |
| GSE77861 | SLAMF9 | 89886 | 1553770_a_at | 0.4963 | 0.0972 | |
| GSE97050 | SLAMF9 | 89886 | A_23_P96833 | -0.1600 | 0.4466 | |
| SRP007169 | SLAMF9 | 89886 | RNAseq | 4.6476 | 0.0009 | |
| TCGA | SLAMF9 | 89886 | RNAseq | 1.5785 | 0.0035 |
Upregulated datasets: 2; Downregulated datasets: 0.
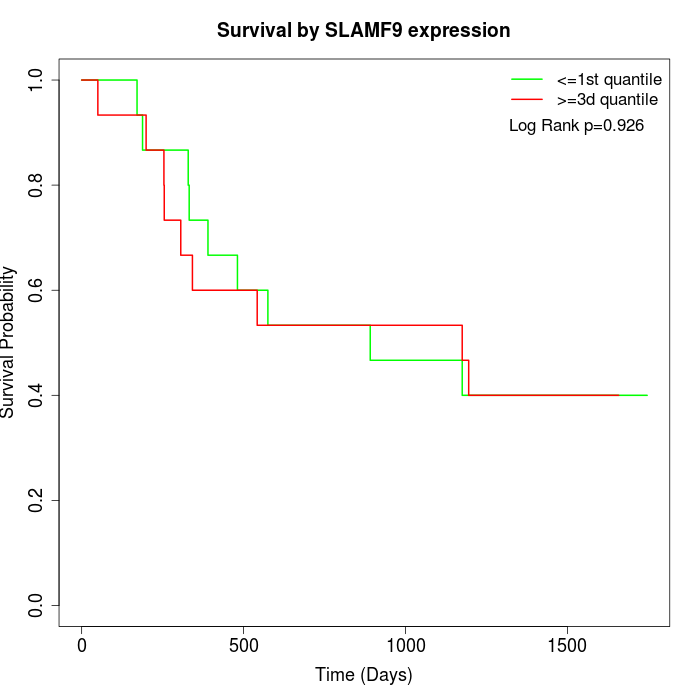
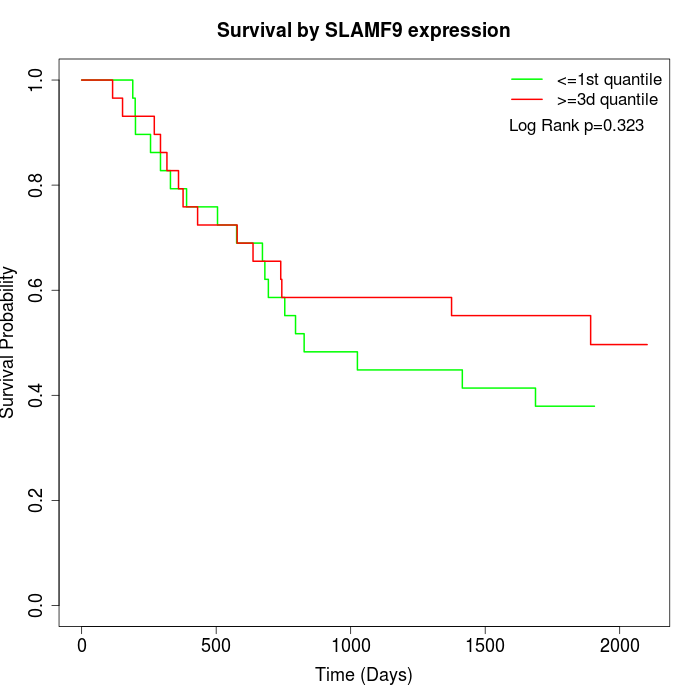
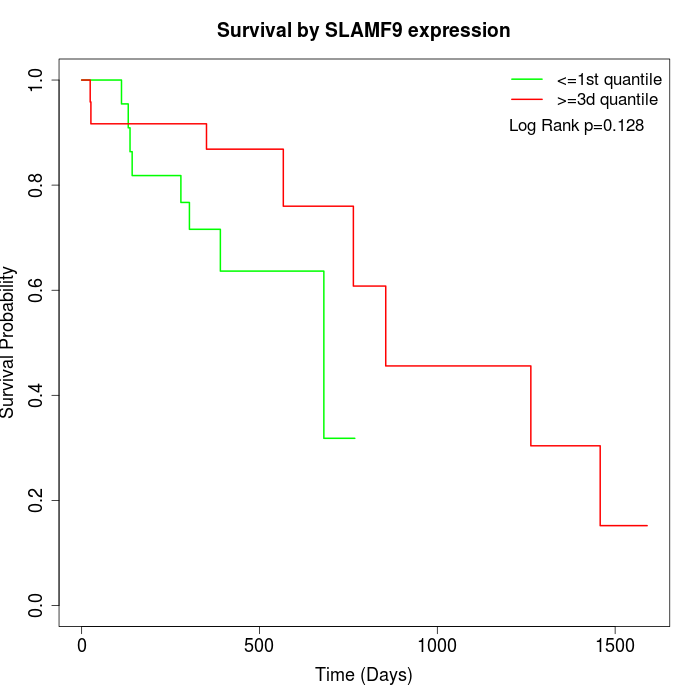
Survival by SLAMF9 expression:
Note: Click image to view full size file.
Copy number change of SLAMF9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SLAMF9 | 89886 | 10 | 0 | 20 | |
| GSE20123 | SLAMF9 | 89886 | 10 | 0 | 20 | |
| GSE43470 | SLAMF9 | 89886 | 7 | 2 | 34 | |
| GSE46452 | SLAMF9 | 89886 | 2 | 1 | 56 | |
| GSE47630 | SLAMF9 | 89886 | 14 | 0 | 26 | |
| GSE54993 | SLAMF9 | 89886 | 0 | 5 | 65 | |
| GSE54994 | SLAMF9 | 89886 | 16 | 0 | 37 | |
| GSE60625 | SLAMF9 | 89886 | 0 | 0 | 11 | |
| GSE74703 | SLAMF9 | 89886 | 7 | 2 | 27 | |
| GSE74704 | SLAMF9 | 89886 | 4 | 0 | 16 | |
| TCGA | SLAMF9 | 89886 | 43 | 3 | 50 |
Total number of gains: 113; Total number of losses: 13; Total Number of normals: 362.
Somatic mutations of SLAMF9:
Generating mutation plots.
Highly correlated genes for SLAMF9:
Showing top 20/215 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SLAMF9 | OR1L6 | 0.792817 | 3 | 0 | 3 |
| SLAMF9 | CALB1 | 0.779995 | 3 | 0 | 3 |
| SLAMF9 | SPEG | 0.753142 | 3 | 0 | 3 |
| SLAMF9 | DUS2 | 0.742032 | 3 | 0 | 3 |
| SLAMF9 | KRTAP10-3 | 0.739865 | 3 | 0 | 3 |
| SLAMF9 | TCF23 | 0.735001 | 3 | 0 | 3 |
| SLAMF9 | TMEM235 | 0.734948 | 3 | 0 | 3 |
| SLAMF9 | RBM17 | 0.723034 | 3 | 0 | 3 |
| SLAMF9 | LEPROTL1 | 0.722554 | 5 | 0 | 5 |
| SLAMF9 | GRIA4 | 0.712179 | 3 | 0 | 3 |
| SLAMF9 | FAM192A | 0.709042 | 4 | 0 | 3 |
| SLAMF9 | MDH1B | 0.707968 | 3 | 0 | 3 |
| SLAMF9 | CDH24 | 0.706013 | 4 | 0 | 4 |
| SLAMF9 | F2RL3 | 0.7049 | 3 | 0 | 3 |
| SLAMF9 | KRTAP10-9 | 0.700837 | 3 | 0 | 3 |
| SLAMF9 | USP2-AS1 | 0.691995 | 3 | 0 | 3 |
| SLAMF9 | FOSL1 | 0.678035 | 3 | 0 | 3 |
| SLAMF9 | MMP2 | 0.677512 | 3 | 0 | 3 |
| SLAMF9 | USP17L2 | 0.671723 | 3 | 0 | 3 |
| SLAMF9 | MSANTD3 | 0.671323 | 3 | 0 | 3 |
For details and further investigation, click here