| Full name: SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Alias Symbol: BAF60A|Rsc6p|CRACD1 | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 6602 | HGNC ID: HGNC:11106 | Ensembl Gene: ENSG00000066117 | OMIM ID: 601735 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SMARCD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMARCD1 | 6602 | 209518_at | 0.4234 | 0.2982 | |
| GSE20347 | SMARCD1 | 6602 | 209518_at | 0.4074 | 0.0002 | |
| GSE23400 | SMARCD1 | 6602 | 209518_at | 0.3169 | 0.0000 | |
| GSE26886 | SMARCD1 | 6602 | 209518_at | 0.4336 | 0.0030 | |
| GSE29001 | SMARCD1 | 6602 | 209518_at | 0.0675 | 0.8778 | |
| GSE38129 | SMARCD1 | 6602 | 209518_at | 0.4304 | 0.0000 | |
| GSE45670 | SMARCD1 | 6602 | 209518_at | 0.1808 | 0.0491 | |
| GSE53622 | SMARCD1 | 6602 | 43339 | 0.7843 | 0.0000 | |
| GSE53624 | SMARCD1 | 6602 | 56882 | 1.0346 | 0.0000 | |
| GSE63941 | SMARCD1 | 6602 | 209518_at | 0.9185 | 0.0035 | |
| GSE77861 | SMARCD1 | 6602 | 209518_at | 0.2255 | 0.2902 | |
| GSE97050 | SMARCD1 | 6602 | A_24_P232696 | 0.4743 | 0.1049 | |
| SRP007169 | SMARCD1 | 6602 | RNAseq | 1.1170 | 0.0033 | |
| SRP008496 | SMARCD1 | 6602 | RNAseq | 0.8587 | 0.0004 | |
| SRP064894 | SMARCD1 | 6602 | RNAseq | 0.4291 | 0.0006 | |
| SRP133303 | SMARCD1 | 6602 | RNAseq | 0.4822 | 0.0000 | |
| SRP159526 | SMARCD1 | 6602 | RNAseq | 0.6951 | 0.0082 | |
| SRP193095 | SMARCD1 | 6602 | RNAseq | 0.5572 | 0.0000 | |
| SRP219564 | SMARCD1 | 6602 | RNAseq | 0.6020 | 0.0324 | |
| TCGA | SMARCD1 | 6602 | RNAseq | 0.2047 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
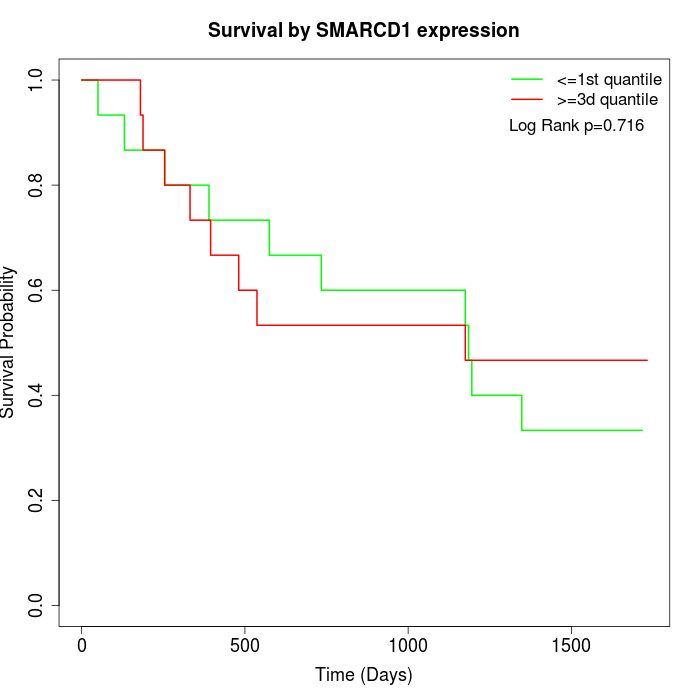
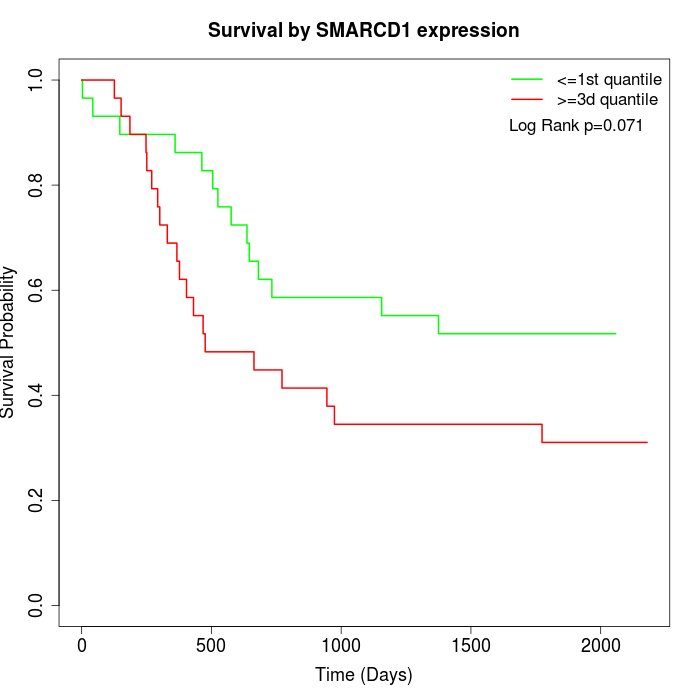
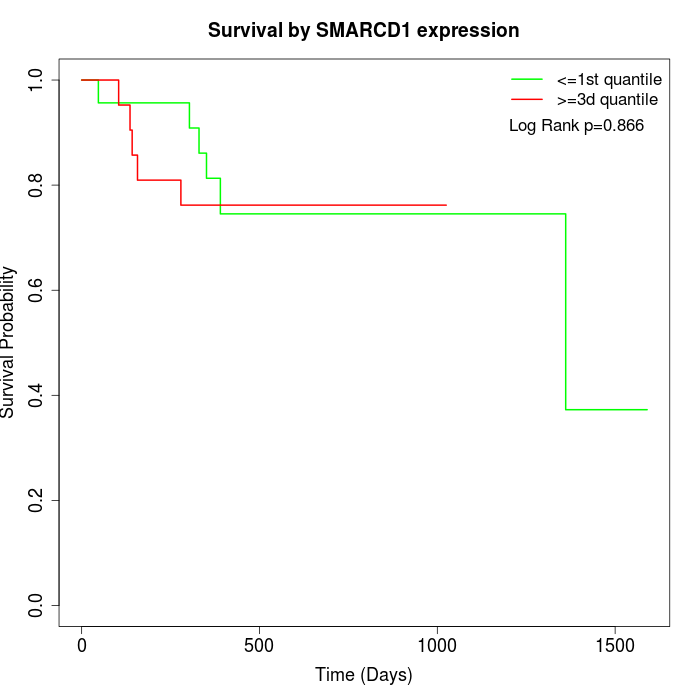
Survival by SMARCD1 expression:
Note: Click image to view full size file.
Copy number change of SMARCD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMARCD1 | 6602 | 8 | 1 | 21 | |
| GSE20123 | SMARCD1 | 6602 | 8 | 1 | 21 | |
| GSE43470 | SMARCD1 | 6602 | 3 | 0 | 40 | |
| GSE46452 | SMARCD1 | 6602 | 8 | 1 | 50 | |
| GSE47630 | SMARCD1 | 6602 | 11 | 2 | 27 | |
| GSE54993 | SMARCD1 | 6602 | 0 | 5 | 65 | |
| GSE54994 | SMARCD1 | 6602 | 4 | 1 | 48 | |
| GSE60625 | SMARCD1 | 6602 | 0 | 0 | 11 | |
| GSE74703 | SMARCD1 | 6602 | 3 | 0 | 33 | |
| GSE74704 | SMARCD1 | 6602 | 5 | 1 | 14 | |
| TCGA | SMARCD1 | 6602 | 15 | 11 | 70 |
Total number of gains: 65; Total number of losses: 23; Total Number of normals: 400.
Somatic mutations of SMARCD1:
Generating mutation plots.
Highly correlated genes for SMARCD1:
Showing top 20/1435 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMARCD1 | FZR1 | 0.813267 | 3 | 0 | 3 |
| SMARCD1 | RFNG | 0.790841 | 3 | 0 | 3 |
| SMARCD1 | GPR160 | 0.788842 | 3 | 0 | 3 |
| SMARCD1 | HIVEP3 | 0.783882 | 3 | 0 | 3 |
| SMARCD1 | NAA60 | 0.775815 | 4 | 0 | 4 |
| SMARCD1 | ZNF707 | 0.764424 | 3 | 0 | 3 |
| SMARCD1 | UCK2 | 0.762455 | 3 | 0 | 3 |
| SMARCD1 | FAM89A | 0.76104 | 3 | 0 | 3 |
| SMARCD1 | NRGN | 0.757372 | 3 | 0 | 3 |
| SMARCD1 | ZNF358 | 0.753271 | 3 | 0 | 3 |
| SMARCD1 | UBE2J2 | 0.746202 | 3 | 0 | 3 |
| SMARCD1 | C11orf68 | 0.740153 | 4 | 0 | 4 |
| SMARCD1 | DOCK5 | 0.739052 | 3 | 0 | 3 |
| SMARCD1 | TMTC4 | 0.738501 | 3 | 0 | 3 |
| SMARCD1 | ADCK1 | 0.738433 | 3 | 0 | 3 |
| SMARCD1 | THOC2 | 0.737243 | 3 | 0 | 3 |
| SMARCD1 | ANGPT2 | 0.735632 | 3 | 0 | 3 |
| SMARCD1 | SCNM1 | 0.735054 | 3 | 0 | 3 |
| SMARCD1 | GGT5 | 0.733064 | 3 | 0 | 3 |
| SMARCD1 | TRAPPC5 | 0.732374 | 3 | 0 | 3 |
For details and further investigation, click here