| Full name: N(alpha)-acetyltransferase 60, NatF catalytic subunit | Alias Symbol: FLJ14154 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 79903 | HGNC ID: HGNC:25875 | Ensembl Gene: ENSG00000122390 | OMIM ID: 614246 |
| Drug and gene relationship at DGIdb | |||
Expression of NAA60:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAA60 | 79903 | 45526_g_at | 0.0879 | 0.7971 | |
| GSE20347 | NAA60 | 79903 | 45526_g_at | -0.0453 | 0.6807 | |
| GSE23400 | NAA60 | 79903 | 45526_g_at | -0.0711 | 0.1313 | |
| GSE26886 | NAA60 | 79903 | 45526_g_at | 0.5670 | 0.0090 | |
| GSE29001 | NAA60 | 79903 | 45526_g_at | -0.0608 | 0.8377 | |
| GSE38129 | NAA60 | 79903 | 45526_g_at | -0.0783 | 0.3806 | |
| GSE45670 | NAA60 | 79903 | 45526_g_at | 0.0145 | 0.9159 | |
| GSE53622 | NAA60 | 79903 | 7831 | -0.2170 | 0.0009 | |
| GSE53624 | NAA60 | 79903 | 7831 | -0.0261 | 0.6765 | |
| GSE63941 | NAA60 | 79903 | 45526_g_at | 0.0768 | 0.7752 | |
| GSE77861 | NAA60 | 79903 | 45526_g_at | -0.1656 | 0.4395 | |
| GSE97050 | NAA60 | 79903 | A_33_P3424267 | 0.3274 | 0.1942 | |
| SRP007169 | NAA60 | 79903 | RNAseq | -1.0859 | 0.0055 | |
| SRP008496 | NAA60 | 79903 | RNAseq | -0.8229 | 0.0008 | |
| SRP064894 | NAA60 | 79903 | RNAseq | -0.1332 | 0.5175 | |
| SRP133303 | NAA60 | 79903 | RNAseq | 0.0829 | 0.4831 | |
| SRP159526 | NAA60 | 79903 | RNAseq | 0.0389 | 0.8948 | |
| SRP193095 | NAA60 | 79903 | RNAseq | -0.1927 | 0.2644 | |
| SRP219564 | NAA60 | 79903 | RNAseq | 0.0872 | 0.7562 |
Upregulated datasets: 0; Downregulated datasets: 1.
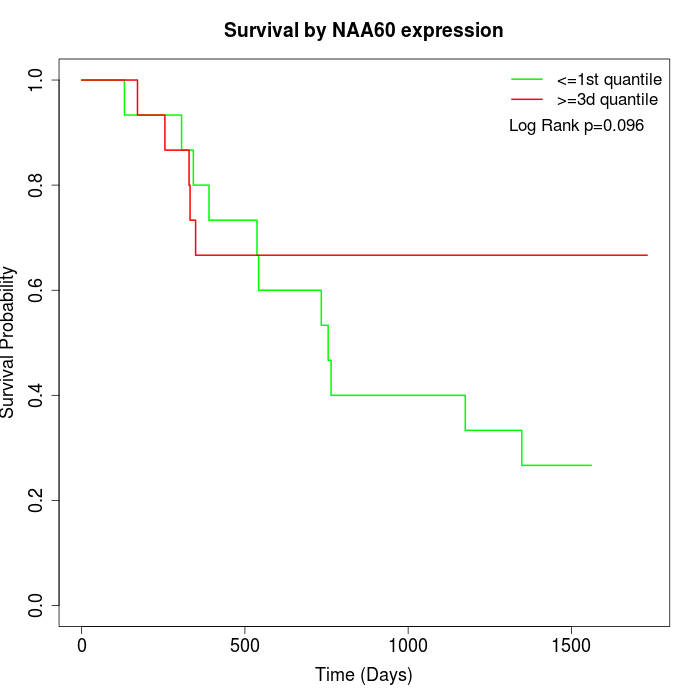
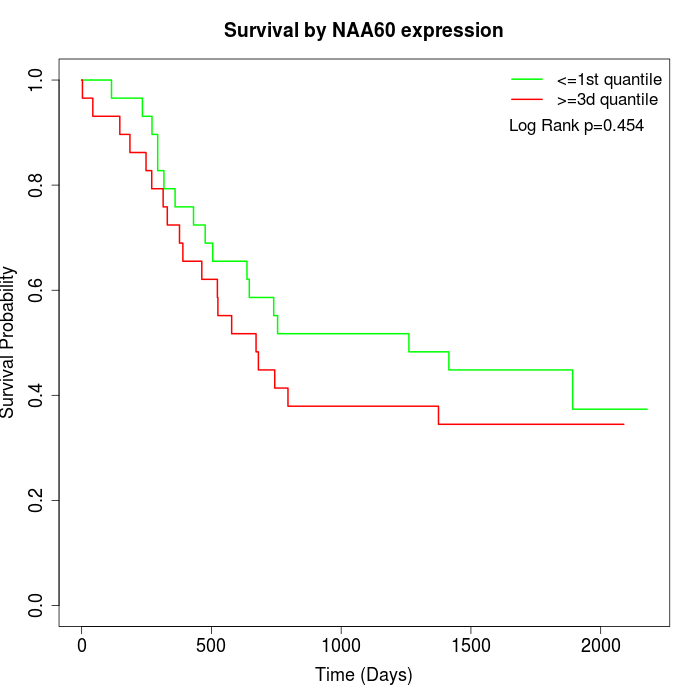
Survival by NAA60 expression:
Note: Click image to view full size file.
Copy number change of NAA60:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAA60 | 79903 | 5 | 5 | 20 | |
| GSE20123 | NAA60 | 79903 | 5 | 4 | 21 | |
| GSE43470 | NAA60 | 79903 | 4 | 7 | 32 | |
| GSE46452 | NAA60 | 79903 | 38 | 1 | 20 | |
| GSE47630 | NAA60 | 79903 | 13 | 6 | 21 | |
| GSE54993 | NAA60 | 79903 | 4 | 5 | 61 | |
| GSE54994 | NAA60 | 79903 | 5 | 9 | 39 | |
| GSE60625 | NAA60 | 79903 | 4 | 0 | 7 | |
| GSE74703 | NAA60 | 79903 | 4 | 5 | 27 | |
| GSE74704 | NAA60 | 79903 | 3 | 2 | 15 | |
| TCGA | NAA60 | 79903 | 19 | 13 | 64 |
Total number of gains: 104; Total number of losses: 57; Total Number of normals: 327.
Somatic mutations of NAA60:
Generating mutation plots.
Highly correlated genes for NAA60:
Showing top 20/267 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAA60 | NACC1 | 0.843103 | 3 | 0 | 3 |
| NAA60 | LUC7L2 | 0.804782 | 3 | 0 | 3 |
| NAA60 | SMARCD1 | 0.775815 | 4 | 0 | 4 |
| NAA60 | CHERP | 0.768527 | 3 | 0 | 3 |
| NAA60 | PRKCSH | 0.761848 | 4 | 0 | 3 |
| NAA60 | POLR2L | 0.750849 | 3 | 0 | 3 |
| NAA60 | MAZ | 0.749745 | 3 | 0 | 3 |
| NAA60 | MRPL4 | 0.748716 | 3 | 0 | 3 |
| NAA60 | CCDC124 | 0.748551 | 3 | 0 | 3 |
| NAA60 | STARD10 | 0.746577 | 3 | 0 | 3 |
| NAA60 | PIN1 | 0.737493 | 3 | 0 | 3 |
| NAA60 | MIER2 | 0.736229 | 3 | 0 | 3 |
| NAA60 | RHBDD2 | 0.728153 | 3 | 0 | 3 |
| NAA60 | SUGP1 | 0.728046 | 4 | 0 | 4 |
| NAA60 | DEDD2 | 0.721601 | 3 | 0 | 3 |
| NAA60 | GCDH | 0.719192 | 4 | 0 | 4 |
| NAA60 | PEX10 | 0.718413 | 4 | 0 | 3 |
| NAA60 | ERCC4 | 0.711428 | 3 | 0 | 3 |
| NAA60 | SHMT2 | 0.711215 | 3 | 0 | 3 |
| NAA60 | C19orf54 | 0.709862 | 4 | 0 | 3 |
For details and further investigation, click here