| Full name: FEV transcription factor, ETS family member | Alias Symbol: Pet-1 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 54738 | HGNC ID: HGNC:18562 | Ensembl Gene: ENSG00000163497 | OMIM ID: 607150 |
| Drug and gene relationship at DGIdb | |||
Expression of FEV:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FEV | 54738 | 207260_at | 0.0211 | 0.9548 | |
| GSE20347 | FEV | 54738 | 207260_at | -0.0511 | 0.6269 | |
| GSE23400 | FEV | 54738 | 207260_at | -0.2060 | 0.0000 | |
| GSE26886 | FEV | 54738 | 207260_at | -0.0012 | 0.9925 | |
| GSE29001 | FEV | 54738 | 207260_at | -0.4800 | 0.0132 | |
| GSE38129 | FEV | 54738 | 207260_at | -0.2272 | 0.0281 | |
| GSE45670 | FEV | 54738 | 207260_at | -0.0223 | 0.8541 | |
| GSE53622 | FEV | 54738 | 1946 | 0.6564 | 0.0010 | |
| GSE53624 | FEV | 54738 | 1946 | 0.4417 | 0.0000 | |
| GSE63941 | FEV | 54738 | 207260_at | 0.3719 | 0.0289 | |
| GSE77861 | FEV | 54738 | 207260_at | -0.1752 | 0.3241 | |
| GSE97050 | FEV | 54738 | A_23_P5460 | -0.1403 | 0.5920 | |
| TCGA | FEV | 54738 | RNAseq | -3.7032 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 1.
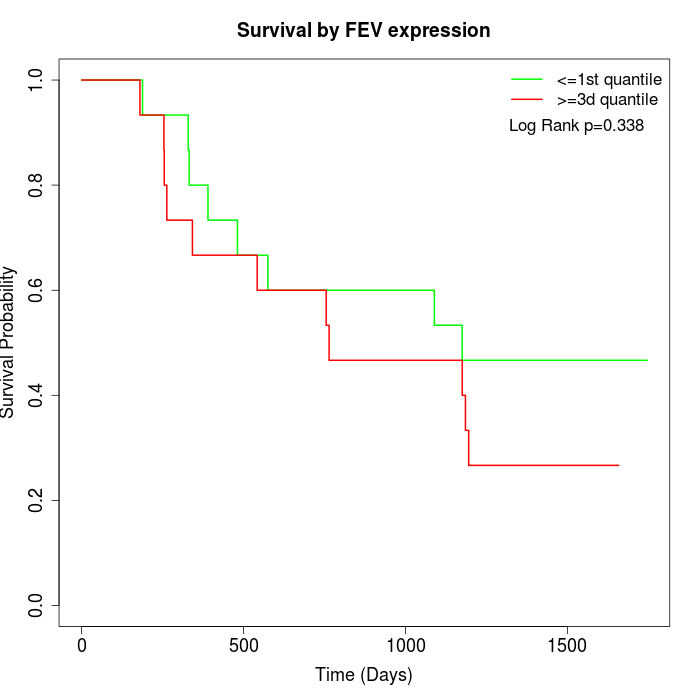
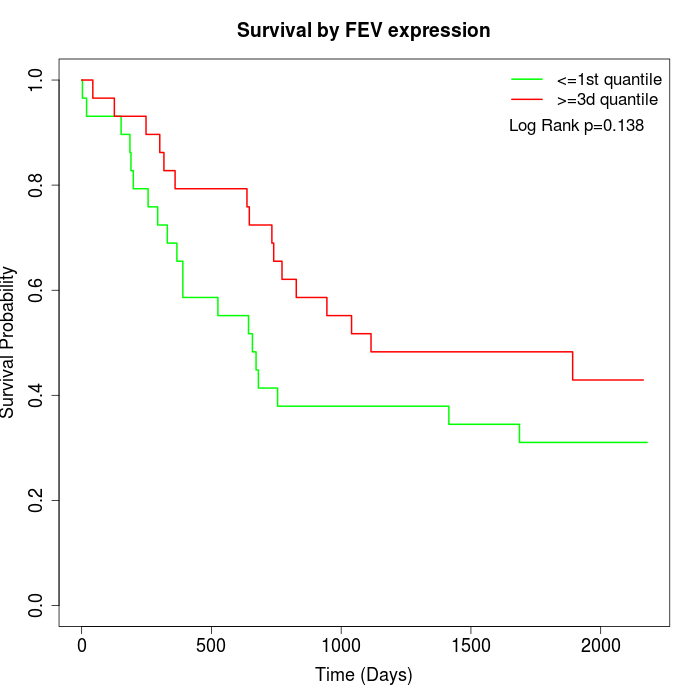
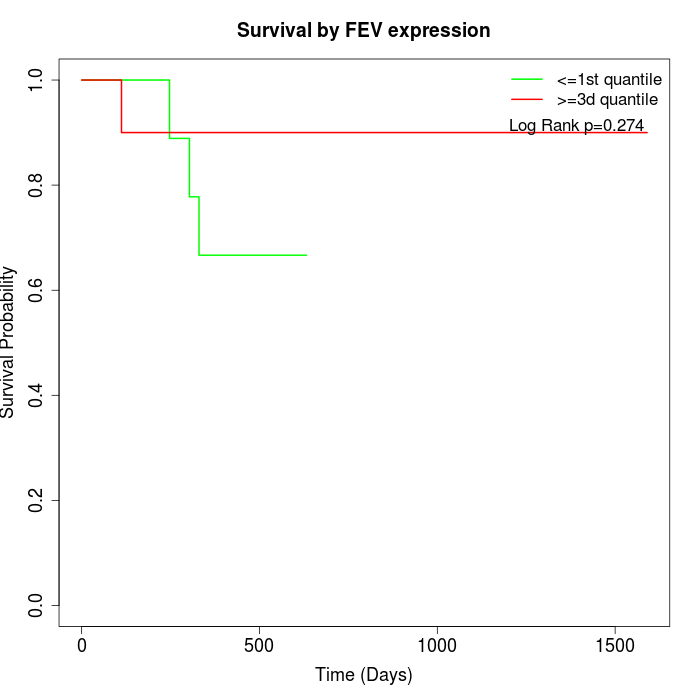
Survival by FEV expression:
Note: Click image to view full size file.
Copy number change of FEV:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FEV | 54738 | 1 | 14 | 15 | |
| GSE20123 | FEV | 54738 | 1 | 13 | 16 | |
| GSE43470 | FEV | 54738 | 1 | 7 | 35 | |
| GSE46452 | FEV | 54738 | 0 | 5 | 54 | |
| GSE47630 | FEV | 54738 | 4 | 5 | 31 | |
| GSE54993 | FEV | 54738 | 1 | 2 | 67 | |
| GSE54994 | FEV | 54738 | 7 | 10 | 36 | |
| GSE60625 | FEV | 54738 | 0 | 3 | 8 | |
| GSE74703 | FEV | 54738 | 1 | 5 | 30 | |
| GSE74704 | FEV | 54738 | 1 | 7 | 12 | |
| TCGA | FEV | 54738 | 11 | 27 | 58 |
Total number of gains: 28; Total number of losses: 98; Total Number of normals: 362.
Somatic mutations of FEV:
Generating mutation plots.
Highly correlated genes for FEV:
Showing top 20/877 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FEV | SERPINA4 | 0.729391 | 6 | 0 | 6 |
| FEV | SEZ6L | 0.715334 | 4 | 0 | 4 |
| FEV | CIDEC | 0.711775 | 5 | 0 | 5 |
| FEV | UBQLN3 | 0.710811 | 4 | 0 | 4 |
| FEV | DNAH11 | 0.702929 | 3 | 0 | 3 |
| FEV | BTNL3 | 0.702628 | 8 | 0 | 8 |
| FEV | DRD5 | 0.695371 | 6 | 0 | 6 |
| FEV | ECE2 | 0.694082 | 5 | 0 | 5 |
| FEV | GABRR2 | 0.691629 | 6 | 0 | 6 |
| FEV | ASIC2 | 0.684387 | 5 | 0 | 5 |
| FEV | NDST4 | 0.684097 | 3 | 0 | 3 |
| FEV | PPEF1 | 0.6835 | 4 | 0 | 4 |
| FEV | MAPK11 | 0.681061 | 4 | 0 | 4 |
| FEV | ABI3 | 0.677458 | 3 | 0 | 3 |
| FEV | KRTAP6-2 | 0.676413 | 3 | 0 | 3 |
| FEV | IFNA17 | 0.671145 | 4 | 0 | 4 |
| FEV | EPN1 | 0.667307 | 6 | 0 | 4 |
| FEV | SPAM1 | 0.667029 | 7 | 0 | 6 |
| FEV | CGA | 0.666912 | 6 | 0 | 5 |
| FEV | GK2 | 0.666456 | 4 | 0 | 4 |
For details and further investigation, click here