| Full name: galectin 14 | Alias Symbol: PPL13|CLC2 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 56891 | HGNC ID: HGNC:30054 | Ensembl Gene: ENSG00000006659 | OMIM ID: 607260 |
| Drug and gene relationship at DGIdb | |||
Expression of LGALS14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LGALS14 | 56891 | 220158_at | -0.1447 | 0.6439 | |
| GSE20347 | LGALS14 | 56891 | 220158_at | -0.0049 | 0.9480 | |
| GSE23400 | LGALS14 | 56891 | 220158_at | -0.1555 | 0.0007 | |
| GSE26886 | LGALS14 | 56891 | 220158_at | -0.0890 | 0.4156 | |
| GSE29001 | LGALS14 | 56891 | 220158_at | -0.0049 | 0.9795 | |
| GSE38129 | LGALS14 | 56891 | 220158_at | -0.0557 | 0.4590 | |
| GSE45670 | LGALS14 | 56891 | 220158_at | -0.0794 | 0.4768 | |
| GSE53622 | LGALS14 | 56891 | 137175 | 0.0173 | 0.9140 | |
| GSE53624 | LGALS14 | 56891 | 137175 | 0.3462 | 0.0002 | |
| GSE63941 | LGALS14 | 56891 | 220158_at | 0.1261 | 0.3057 | |
| GSE77861 | LGALS14 | 56891 | 220158_at | -0.1485 | 0.1776 | |
| SRP133303 | LGALS14 | 56891 | RNAseq | -0.0486 | 0.7770 |
Upregulated datasets: 0; Downregulated datasets: 0.
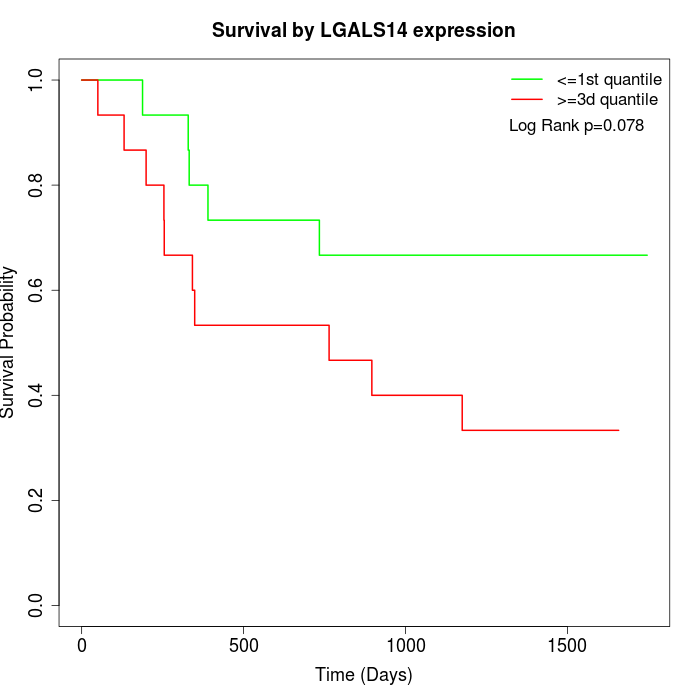
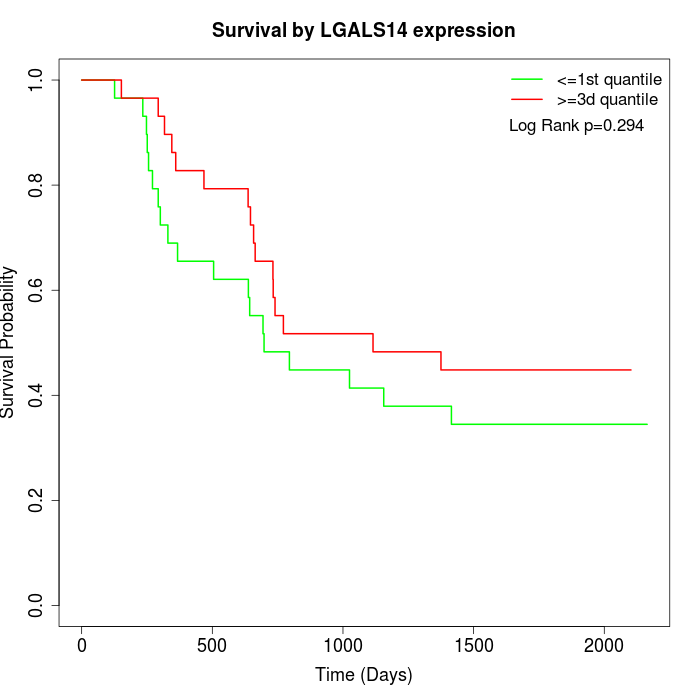
Survival by LGALS14 expression:
Note: Click image to view full size file.
Copy number change of LGALS14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LGALS14 | 56891 | 7 | 4 | 19 | |
| GSE20123 | LGALS14 | 56891 | 7 | 3 | 20 | |
| GSE43470 | LGALS14 | 56891 | 3 | 11 | 29 | |
| GSE46452 | LGALS14 | 56891 | 48 | 1 | 10 | |
| GSE47630 | LGALS14 | 56891 | 9 | 5 | 26 | |
| GSE54993 | LGALS14 | 56891 | 17 | 3 | 50 | |
| GSE54994 | LGALS14 | 56891 | 7 | 9 | 37 | |
| GSE60625 | LGALS14 | 56891 | 9 | 0 | 2 | |
| GSE74703 | LGALS14 | 56891 | 3 | 7 | 26 | |
| GSE74704 | LGALS14 | 56891 | 7 | 1 | 12 | |
| TCGA | LGALS14 | 56891 | 14 | 16 | 66 |
Total number of gains: 131; Total number of losses: 60; Total Number of normals: 297.
Somatic mutations of LGALS14:
Generating mutation plots.
Highly correlated genes for LGALS14:
Showing top 20/801 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LGALS14 | SLC6A5 | 0.783016 | 3 | 0 | 3 |
| LGALS14 | MYH8 | 0.739055 | 3 | 0 | 3 |
| LGALS14 | RHAG | 0.729825 | 3 | 0 | 3 |
| LGALS14 | MTAP | 0.72341 | 4 | 0 | 4 |
| LGALS14 | NOS1 | 0.720396 | 3 | 0 | 3 |
| LGALS14 | ADCYAP1 | 0.717494 | 3 | 0 | 3 |
| LGALS14 | ADCYAP1R1 | 0.708906 | 3 | 0 | 3 |
| LGALS14 | NAT2 | 0.705938 | 3 | 0 | 3 |
| LGALS14 | NCR1 | 0.702959 | 4 | 0 | 4 |
| LGALS14 | CRHR2 | 0.699842 | 4 | 0 | 4 |
| LGALS14 | HRG | 0.690048 | 4 | 0 | 4 |
| LGALS14 | MSI1 | 0.683159 | 3 | 0 | 3 |
| LGALS14 | KIR3DL3 | 0.673633 | 5 | 0 | 4 |
| LGALS14 | SPAM1 | 0.672523 | 5 | 0 | 5 |
| LGALS14 | ABCB5 | 0.668988 | 3 | 0 | 3 |
| LGALS14 | OR3A2 | 0.668659 | 3 | 0 | 3 |
| LGALS14 | ANGPT4 | 0.66791 | 7 | 0 | 6 |
| LGALS14 | FOLH1 | 0.665529 | 6 | 0 | 5 |
| LGALS14 | GOLGA6L6 | 0.664704 | 3 | 0 | 3 |
| LGALS14 | KCNJ10 | 0.663811 | 4 | 0 | 3 |
For details and further investigation, click here