| Full name: TATA-box binding protein associated factor 1 like | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9p21.1 | ||
| Entrez ID: 138474 | HGNC ID: HGNC:18056 | Ensembl Gene: ENSG00000122728 | OMIM ID: 607798 |
| Drug and gene relationship at DGIdb | |||
Expression of TAF1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAF1L | 138474 | 1553011_at | -0.0464 | 0.9009 | |
| GSE26886 | TAF1L | 138474 | 1553011_at | 0.1716 | 0.2419 | |
| GSE45670 | TAF1L | 138474 | 1553011_at | 0.0985 | 0.2695 | |
| GSE53622 | TAF1L | 138474 | 74860 | 0.1057 | 0.1428 | |
| GSE53624 | TAF1L | 138474 | 74860 | 0.2399 | 0.0035 | |
| GSE63941 | TAF1L | 138474 | 1553011_at | 0.1608 | 0.5157 | |
| GSE77861 | TAF1L | 138474 | 1553011_at | -0.0397 | 0.7611 | |
| GSE97050 | TAF1L | 138474 | A_33_P3257457 | -0.1796 | 0.3543 | |
| SRP133303 | TAF1L | 138474 | RNAseq | 0.4508 | 0.1564 | |
| TCGA | TAF1L | 138474 | RNAseq | -0.0746 | 0.5503 |
Upregulated datasets: 0; Downregulated datasets: 0.
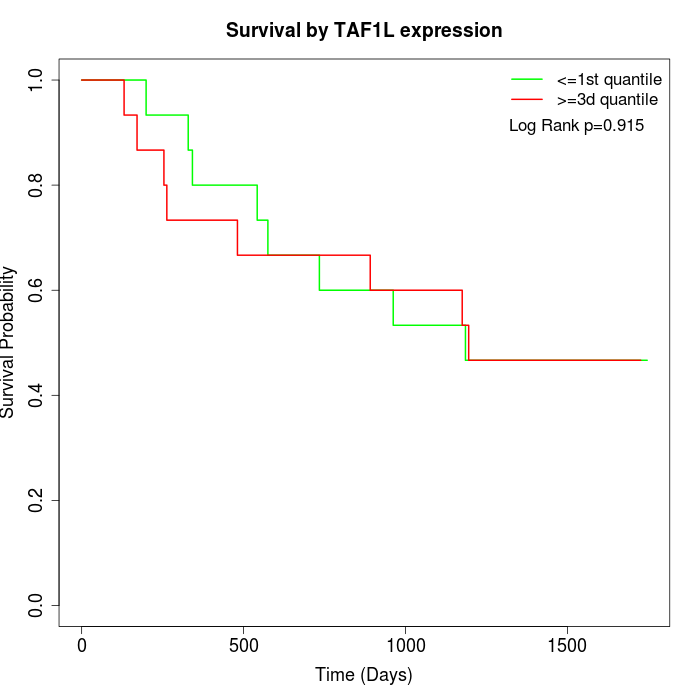
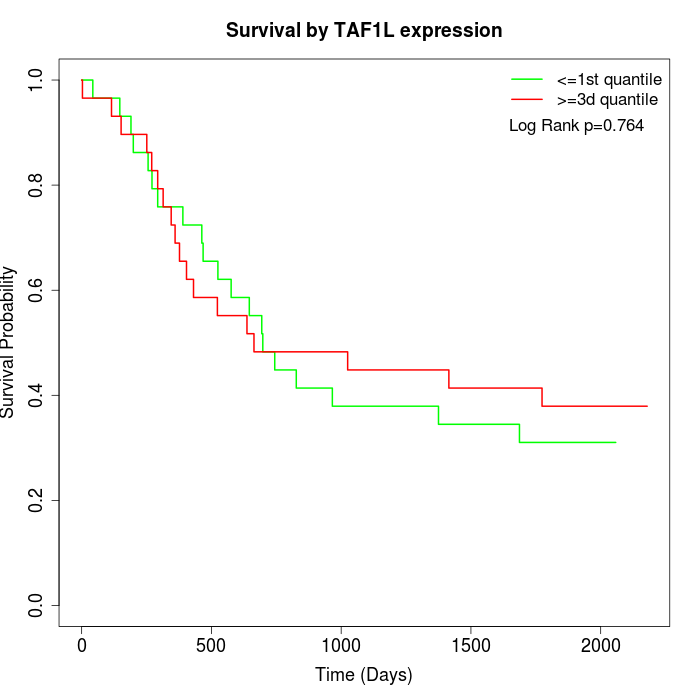
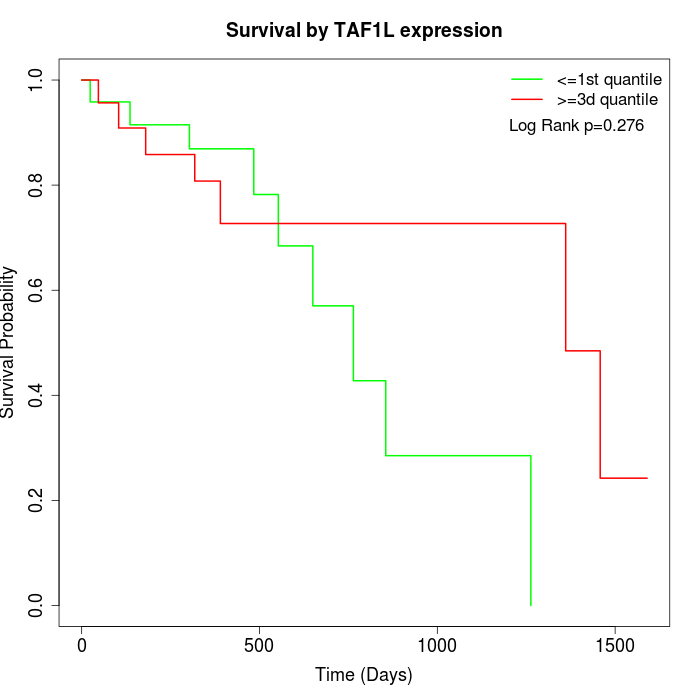
Survival by TAF1L expression:
Note: Click image to view full size file.
Copy number change of TAF1L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAF1L | 138474 | 2 | 15 | 13 | |
| GSE20123 | TAF1L | 138474 | 2 | 15 | 13 | |
| GSE43470 | TAF1L | 138474 | 4 | 9 | 30 | |
| GSE46452 | TAF1L | 138474 | 7 | 15 | 37 | |
| GSE47630 | TAF1L | 138474 | 1 | 21 | 18 | |
| GSE54993 | TAF1L | 138474 | 6 | 0 | 64 | |
| GSE54994 | TAF1L | 138474 | 7 | 11 | 35 | |
| GSE60625 | TAF1L | 138474 | 0 | 0 | 11 | |
| GSE74703 | TAF1L | 138474 | 3 | 7 | 26 | |
| GSE74704 | TAF1L | 138474 | 0 | 11 | 9 | |
| TCGA | TAF1L | 138474 | 16 | 44 | 36 |
Total number of gains: 48; Total number of losses: 148; Total Number of normals: 292.
Somatic mutations of TAF1L:
Generating mutation plots.
Highly correlated genes for TAF1L:
Showing top 20/65 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAF1L | BHLHE41 | 0.762722 | 3 | 0 | 3 |
| TAF1L | PEG10 | 0.735683 | 3 | 0 | 3 |
| TAF1L | SLC13A2 | 0.728628 | 3 | 0 | 3 |
| TAF1L | ADAM32 | 0.7266 | 3 | 0 | 3 |
| TAF1L | SETD5 | 0.721018 | 3 | 0 | 3 |
| TAF1L | CTPS2 | 0.710318 | 3 | 0 | 3 |
| TAF1L | MKS1 | 0.700742 | 3 | 0 | 3 |
| TAF1L | CASQ2 | 0.69703 | 3 | 0 | 3 |
| TAF1L | ACRBP | 0.695812 | 3 | 0 | 3 |
| TAF1L | WIPF3 | 0.693708 | 3 | 0 | 3 |
| TAF1L | ID3 | 0.689506 | 3 | 0 | 3 |
| TAF1L | C10orf82 | 0.674733 | 3 | 0 | 3 |
| TAF1L | NHS | 0.669308 | 3 | 0 | 3 |
| TAF1L | HOXB3 | 0.660902 | 4 | 0 | 3 |
| TAF1L | PPIE | 0.660308 | 3 | 0 | 3 |
| TAF1L | ASB12 | 0.656612 | 4 | 0 | 4 |
| TAF1L | KCNA2 | 0.654934 | 3 | 0 | 3 |
| TAF1L | STOML2 | 0.654394 | 3 | 0 | 3 |
| TAF1L | NEDD4 | 0.653297 | 3 | 0 | 3 |
| TAF1L | SNUPN | 0.652579 | 3 | 0 | 3 |
For details and further investigation, click here