| Full name: translocation associated membrane protein 1 like 1 | Alias Symbol: MGC26568 | ||
| Type: protein-coding gene | Cytoband: 4q26 | ||
| Entrez ID: 133022 | HGNC ID: HGNC:28371 | Ensembl Gene: ENSG00000174599 | OMIM ID: 617505 |
| Drug and gene relationship at DGIdb | |||
Expression of TRAM1L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRAM1L1 | 133022 | 244334_at | 0.2166 | 0.7822 | |
| GSE26886 | TRAM1L1 | 133022 | 244334_at | 0.4794 | 0.2376 | |
| GSE45670 | TRAM1L1 | 133022 | 244334_at | -0.3909 | 0.0393 | |
| GSE53622 | TRAM1L1 | 133022 | 76780 | -0.0677 | 0.6514 | |
| GSE53624 | TRAM1L1 | 133022 | 76780 | 0.0294 | 0.8322 | |
| GSE63941 | TRAM1L1 | 133022 | 244334_at | -2.0604 | 0.0211 | |
| GSE77861 | TRAM1L1 | 133022 | 244334_at | 0.0473 | 0.7998 | |
| GSE97050 | TRAM1L1 | 133022 | A_23_P18518 | -0.0235 | 0.9341 | |
| SRP133303 | TRAM1L1 | 133022 | RNAseq | -0.1421 | 0.7441 | |
| SRP159526 | TRAM1L1 | 133022 | RNAseq | 0.2454 | 0.7398 | |
| SRP219564 | TRAM1L1 | 133022 | RNAseq | -0.8463 | 0.2308 | |
| TCGA | TRAM1L1 | 133022 | RNAseq | -0.6304 | 0.0140 |
Upregulated datasets: 0; Downregulated datasets: 1.
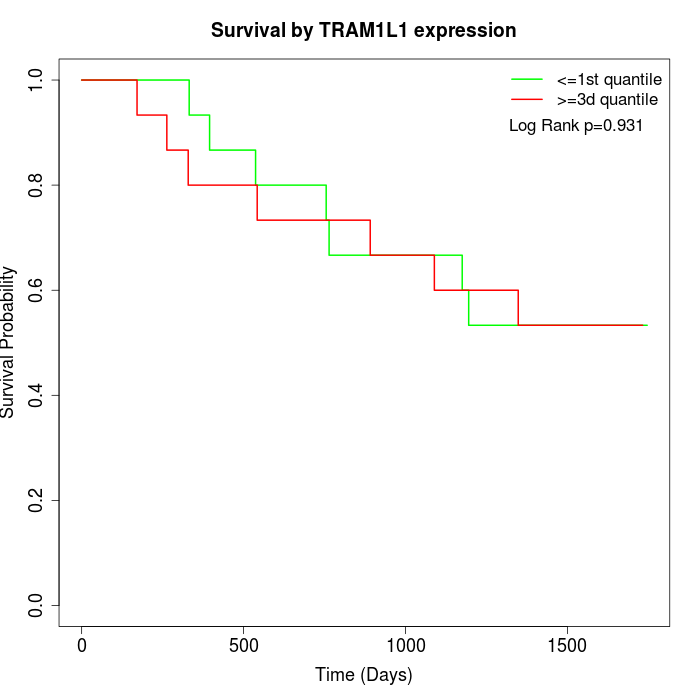
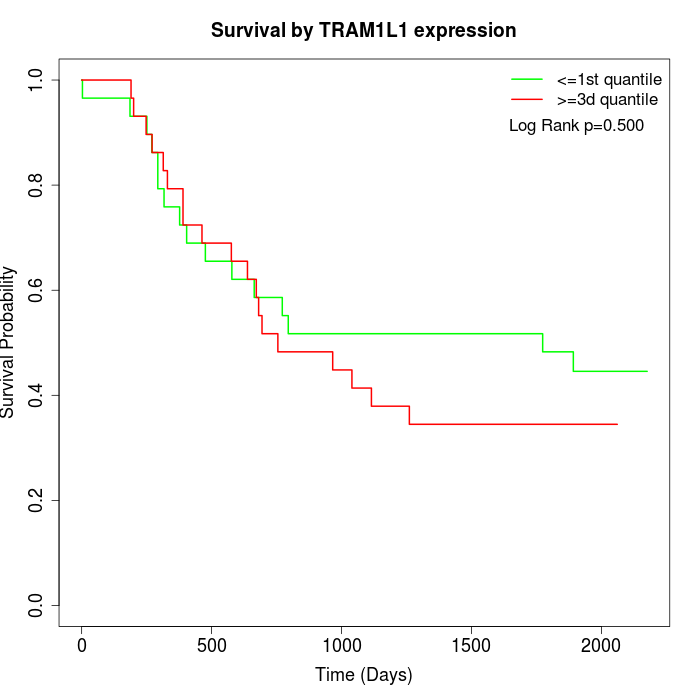
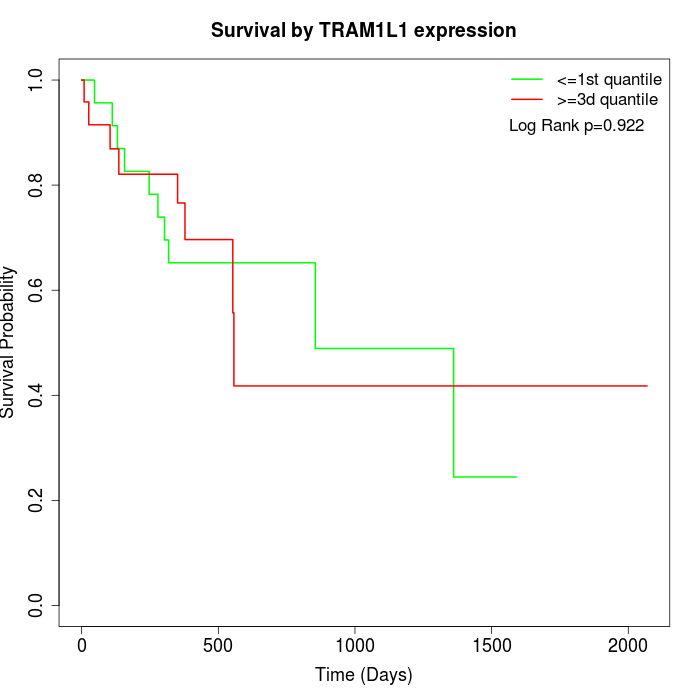
Survival by TRAM1L1 expression:
Note: Click image to view full size file.
Copy number change of TRAM1L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRAM1L1 | 133022 | 0 | 13 | 17 | |
| GSE20123 | TRAM1L1 | 133022 | 1 | 13 | 16 | |
| GSE43470 | TRAM1L1 | 133022 | 0 | 14 | 29 | |
| GSE46452 | TRAM1L1 | 133022 | 1 | 36 | 22 | |
| GSE47630 | TRAM1L1 | 133022 | 0 | 22 | 18 | |
| GSE54993 | TRAM1L1 | 133022 | 9 | 0 | 61 | |
| GSE54994 | TRAM1L1 | 133022 | 1 | 10 | 42 | |
| GSE60625 | TRAM1L1 | 133022 | 0 | 1 | 10 | |
| GSE74703 | TRAM1L1 | 133022 | 0 | 12 | 24 | |
| GSE74704 | TRAM1L1 | 133022 | 0 | 7 | 13 | |
| TCGA | TRAM1L1 | 133022 | 7 | 35 | 54 |
Total number of gains: 19; Total number of losses: 163; Total Number of normals: 306.
Somatic mutations of TRAM1L1:
Generating mutation plots.
Highly correlated genes for TRAM1L1:
Showing top 20/46 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRAM1L1 | CLDN3 | 0.712967 | 3 | 0 | 3 |
| TRAM1L1 | MYBPC1 | 0.681757 | 3 | 0 | 3 |
| TRAM1L1 | DNAL4 | 0.67384 | 4 | 0 | 3 |
| TRAM1L1 | TRAPPC9 | 0.673068 | 3 | 0 | 3 |
| TRAM1L1 | KIAA1211 | 0.671509 | 3 | 0 | 3 |
| TRAM1L1 | ATP7B | 0.66541 | 4 | 0 | 3 |
| TRAM1L1 | TUSC1 | 0.662575 | 3 | 0 | 3 |
| TRAM1L1 | ZNF678 | 0.652953 | 3 | 0 | 3 |
| TRAM1L1 | S1PR2 | 0.64586 | 3 | 0 | 3 |
| TRAM1L1 | SCRN2 | 0.634429 | 3 | 0 | 3 |
| TRAM1L1 | HCK | 0.632982 | 3 | 0 | 3 |
| TRAM1L1 | HEBP1 | 0.630397 | 4 | 0 | 3 |
| TRAM1L1 | CHST14 | 0.620792 | 3 | 0 | 3 |
| TRAM1L1 | CHSY3 | 0.616493 | 3 | 0 | 3 |
| TRAM1L1 | NCAM2 | 0.61257 | 3 | 0 | 3 |
| TRAM1L1 | ZNF48 | 0.612059 | 4 | 0 | 3 |
| TRAM1L1 | DACH1 | 0.606666 | 5 | 0 | 3 |
| TRAM1L1 | PABPC4L | 0.606277 | 5 | 0 | 5 |
| TRAM1L1 | USP27X | 0.604415 | 4 | 0 | 3 |
| TRAM1L1 | FAM171A1 | 0.598927 | 5 | 0 | 3 |
For details and further investigation, click here