| Full name: HCK proto-oncogene, Src family tyrosine kinase | Alias Symbol: JTK9 | ||
| Type: protein-coding gene | Cytoband: 20q11.21 | ||
| Entrez ID: 3055 | HGNC ID: HGNC:4840 | Ensembl Gene: ENSG00000101336 | OMIM ID: 142370 |
| Drug and gene relationship at DGIdb | |||
HCK involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04666 | Fc gamma R-mediated phagocytosis |
Expression of HCK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCK | 3055 | 208018_s_at | 0.7265 | 0.5043 | |
| GSE20347 | HCK | 3055 | 208018_s_at | 0.0951 | 0.5098 | |
| GSE23400 | HCK | 3055 | 208018_s_at | 0.1589 | 0.0832 | |
| GSE26886 | HCK | 3055 | 208018_s_at | 0.0864 | 0.8722 | |
| GSE29001 | HCK | 3055 | 208018_s_at | 0.3854 | 0.1132 | |
| GSE38129 | HCK | 3055 | 208018_s_at | 0.3842 | 0.0145 | |
| GSE45670 | HCK | 3055 | 208018_s_at | 0.4551 | 0.0258 | |
| GSE53622 | HCK | 3055 | 148289 | 0.3600 | 0.0000 | |
| GSE53624 | HCK | 3055 | 148289 | 0.0371 | 0.6194 | |
| GSE63941 | HCK | 3055 | 208018_s_at | 1.3353 | 0.3372 | |
| GSE77861 | HCK | 3055 | 208018_s_at | 0.4232 | 0.1240 | |
| GSE97050 | HCK | 3055 | A_24_P365526 | 0.2891 | 0.2290 | |
| SRP007169 | HCK | 3055 | RNAseq | 2.1484 | 0.0019 | |
| SRP008496 | HCK | 3055 | RNAseq | 1.6912 | 0.0007 | |
| SRP064894 | HCK | 3055 | RNAseq | 1.2039 | 0.0000 | |
| SRP133303 | HCK | 3055 | RNAseq | 1.0616 | 0.0000 | |
| SRP159526 | HCK | 3055 | RNAseq | 0.8828 | 0.1820 | |
| SRP193095 | HCK | 3055 | RNAseq | 1.2542 | 0.0000 | |
| SRP219564 | HCK | 3055 | RNAseq | 0.9745 | 0.0852 | |
| TCGA | HCK | 3055 | RNAseq | 0.5876 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
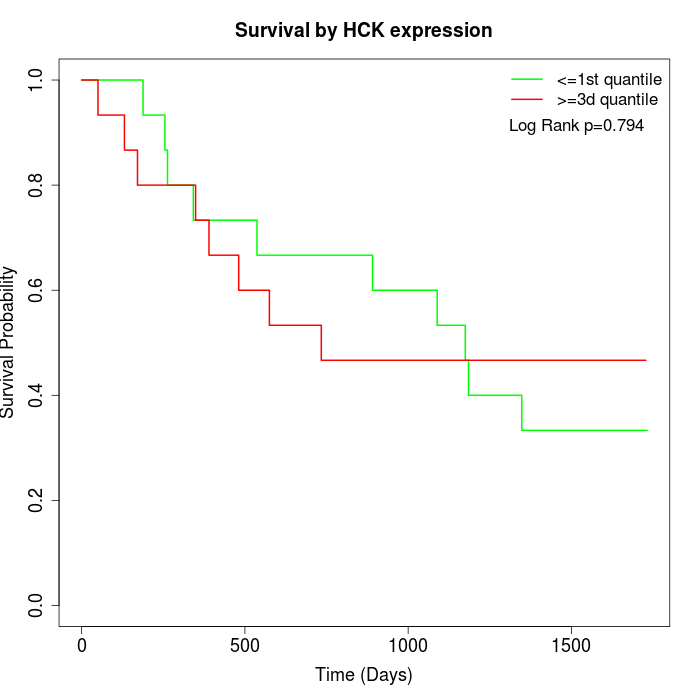
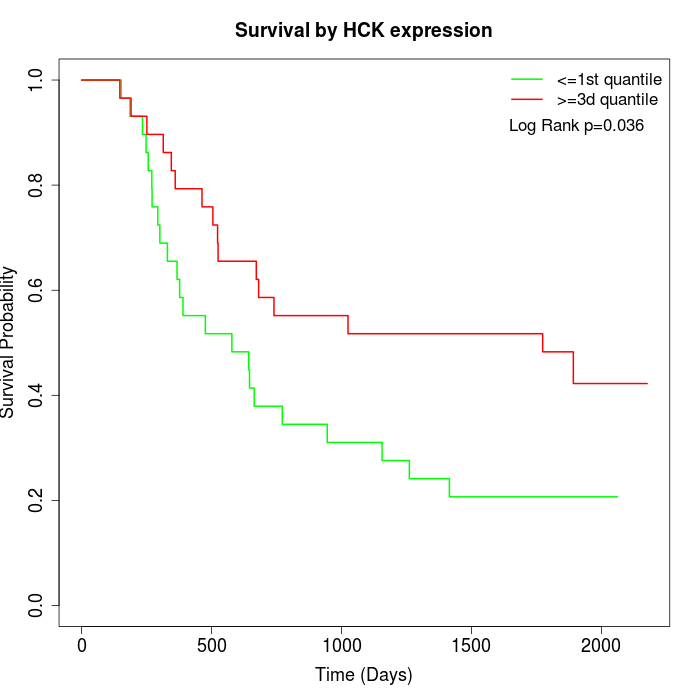
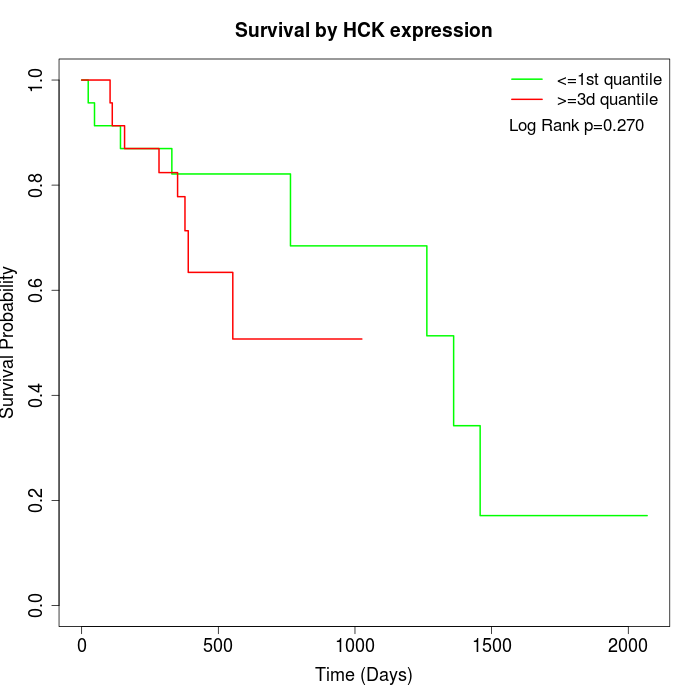
Survival by HCK expression:
Note: Click image to view full size file.
Copy number change of HCK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCK | 3055 | 15 | 0 | 15 | |
| GSE20123 | HCK | 3055 | 15 | 0 | 15 | |
| GSE43470 | HCK | 3055 | 11 | 0 | 32 | |
| GSE46452 | HCK | 3055 | 29 | 0 | 30 | |
| GSE47630 | HCK | 3055 | 25 | 0 | 15 | |
| GSE54993 | HCK | 3055 | 0 | 18 | 52 | |
| GSE54994 | HCK | 3055 | 27 | 1 | 25 | |
| GSE60625 | HCK | 3055 | 0 | 0 | 11 | |
| GSE74703 | HCK | 3055 | 10 | 0 | 26 | |
| GSE74704 | HCK | 3055 | 10 | 0 | 10 | |
| TCGA | HCK | 3055 | 49 | 1 | 46 |
Total number of gains: 191; Total number of losses: 20; Total Number of normals: 277.
Somatic mutations of HCK:
Generating mutation plots.
Highly correlated genes for HCK:
Showing top 20/151 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCK | METTL7B | 0.724493 | 3 | 0 | 3 |
| HCK | TBC1D10C | 0.707461 | 3 | 0 | 3 |
| HCK | ACP5 | 0.681376 | 4 | 0 | 3 |
| HCK | PRAME | 0.679165 | 5 | 0 | 4 |
| HCK | POM121L12 | 0.674051 | 3 | 0 | 3 |
| HCK | SHC2 | 0.669395 | 3 | 0 | 3 |
| HCK | SLC5A6 | 0.66877 | 3 | 0 | 3 |
| HCK | PCED1B | 0.665612 | 3 | 0 | 3 |
| HCK | ABI3 | 0.65875 | 4 | 0 | 3 |
| HCK | MEI1 | 0.65698 | 4 | 0 | 3 |
| HCK | FFAR2 | 0.654411 | 4 | 0 | 3 |
| HCK | ZFAT | 0.654148 | 3 | 0 | 3 |
| HCK | ADAMTS2 | 0.653314 | 4 | 0 | 4 |
| HCK | TSPAN11 | 0.644716 | 5 | 0 | 4 |
| HCK | GCLM | 0.644575 | 4 | 0 | 3 |
| HCK | CRABP1 | 0.642769 | 3 | 0 | 3 |
| HCK | HAGHL | 0.634218 | 4 | 0 | 3 |
| HCK | SKAP1 | 0.634144 | 4 | 0 | 3 |
| HCK | TRAM1L1 | 0.632982 | 3 | 0 | 3 |
| HCK | UBE2I | 0.624936 | 6 | 0 | 5 |
For details and further investigation, click here