| Full name: tripartite motif containing 22 | Alias Symbol: STAF50|GPSTAF50|RNF94 | ||
| Type: protein-coding gene | Cytoband: 11p15.4 | ||
| Entrez ID: 10346 | HGNC ID: HGNC:16379 | Ensembl Gene: ENSG00000132274 | OMIM ID: 606559 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM22 | 10346 | 213293_s_at | -0.5033 | 0.6135 | |
| GSE20347 | TRIM22 | 10346 | 213293_s_at | -0.2990 | 0.4670 | |
| GSE23400 | TRIM22 | 10346 | 213293_s_at | 0.2877 | 0.1357 | |
| GSE26886 | TRIM22 | 10346 | 213293_s_at | -0.1780 | 0.7654 | |
| GSE29001 | TRIM22 | 10346 | 213293_s_at | 0.4829 | 0.2858 | |
| GSE38129 | TRIM22 | 10346 | 213293_s_at | -0.4360 | 0.1080 | |
| GSE45670 | TRIM22 | 10346 | 213293_s_at | 0.2230 | 0.6748 | |
| GSE53622 | TRIM22 | 10346 | 76781 | -0.0430 | 0.7692 | |
| GSE53624 | TRIM22 | 10346 | 76781 | -0.1026 | 0.4549 | |
| GSE63941 | TRIM22 | 10346 | 213293_s_at | -4.4468 | 0.0338 | |
| GSE77861 | TRIM22 | 10346 | 213293_s_at | -1.2827 | 0.0179 | |
| GSE97050 | TRIM22 | 10346 | A_24_P172481 | 0.4030 | 0.3883 | |
| SRP007169 | TRIM22 | 10346 | RNAseq | 0.4314 | 0.2828 | |
| SRP008496 | TRIM22 | 10346 | RNAseq | 0.1898 | 0.4964 | |
| SRP064894 | TRIM22 | 10346 | RNAseq | 0.5876 | 0.0873 | |
| SRP133303 | TRIM22 | 10346 | RNAseq | -0.0213 | 0.9252 | |
| SRP159526 | TRIM22 | 10346 | RNAseq | -1.2019 | 0.0506 | |
| SRP193095 | TRIM22 | 10346 | RNAseq | 0.2391 | 0.1931 | |
| SRP219564 | TRIM22 | 10346 | RNAseq | 0.4143 | 0.4413 | |
| TCGA | TRIM22 | 10346 | RNAseq | 0.0843 | 0.4471 |
Upregulated datasets: 0; Downregulated datasets: 2.
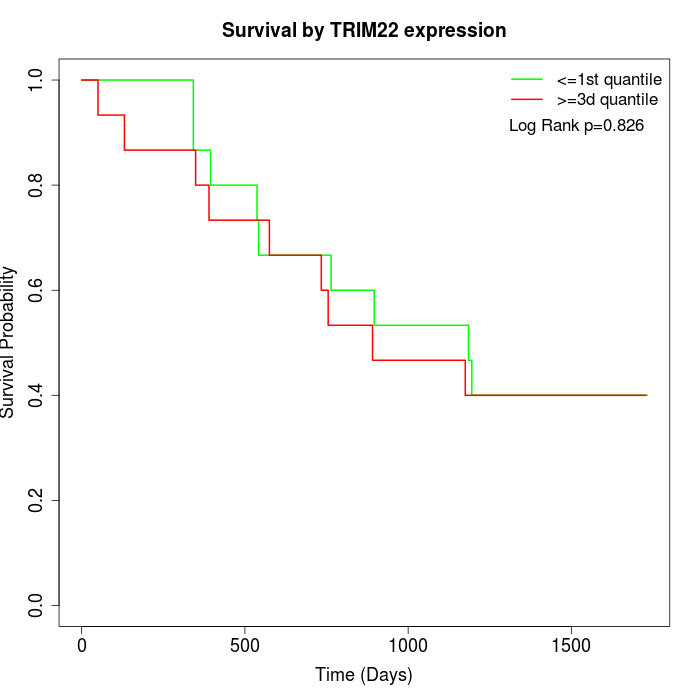
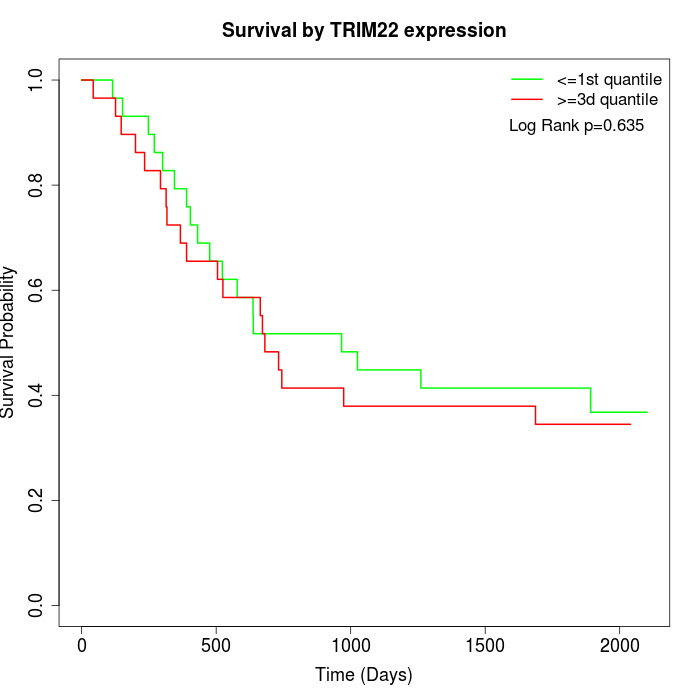
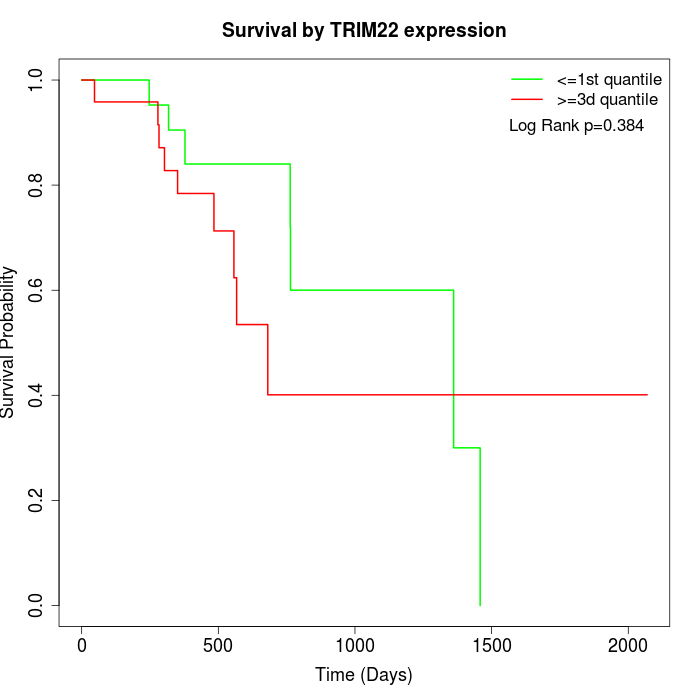
Survival by TRIM22 expression:
Note: Click image to view full size file.
Copy number change of TRIM22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM22 | 10346 | 1 | 11 | 18 | |
| GSE20123 | TRIM22 | 10346 | 1 | 11 | 18 | |
| GSE43470 | TRIM22 | 10346 | 1 | 8 | 34 | |
| GSE46452 | TRIM22 | 10346 | 7 | 8 | 44 | |
| GSE47630 | TRIM22 | 10346 | 3 | 13 | 24 | |
| GSE54993 | TRIM22 | 10346 | 3 | 1 | 66 | |
| GSE54994 | TRIM22 | 10346 | 2 | 12 | 39 | |
| GSE60625 | TRIM22 | 10346 | 0 | 0 | 11 | |
| GSE74703 | TRIM22 | 10346 | 1 | 6 | 29 | |
| GSE74704 | TRIM22 | 10346 | 0 | 8 | 12 | |
| TCGA | TRIM22 | 10346 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 117; Total Number of normals: 344.
Somatic mutations of TRIM22:
Generating mutation plots.
Highly correlated genes for TRIM22:
Showing top 20/421 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM22 | LRRK2 | 0.740179 | 4 | 0 | 4 |
| TRIM22 | UFL1 | 0.736121 | 3 | 0 | 3 |
| TRIM22 | ZNF33A | 0.732334 | 3 | 0 | 3 |
| TRIM22 | NOTCH2 | 0.731643 | 3 | 0 | 3 |
| TRIM22 | RWDD2A | 0.724319 | 3 | 0 | 3 |
| TRIM22 | FLT3LG | 0.723975 | 3 | 0 | 3 |
| TRIM22 | SAMD9L | 0.721561 | 9 | 0 | 9 |
| TRIM22 | APOOL | 0.721242 | 3 | 0 | 3 |
| TRIM22 | MACROD2 | 0.719468 | 3 | 0 | 3 |
| TRIM22 | RPL41 | 0.716515 | 3 | 0 | 3 |
| TRIM22 | RBM20 | 0.714466 | 3 | 0 | 3 |
| TRIM22 | CNTNAP1 | 0.714058 | 3 | 0 | 3 |
| TRIM22 | PDLIM5 | 0.713338 | 3 | 0 | 3 |
| TRIM22 | ZNF441 | 0.709858 | 4 | 0 | 4 |
| TRIM22 | UCHL3 | 0.706675 | 3 | 0 | 3 |
| TRIM22 | TMEM51 | 0.698332 | 3 | 0 | 3 |
| TRIM22 | COMMD3 | 0.695474 | 3 | 0 | 3 |
| TRIM22 | SGMS2 | 0.694045 | 4 | 0 | 4 |
| TRIM22 | SLC35B3 | 0.69022 | 3 | 0 | 3 |
| TRIM22 | NBR1 | 0.68917 | 3 | 0 | 3 |
For details and further investigation, click here