| Full name: VAC14 component of PIKFYVE complex | Alias Symbol: FLJ10305|ArPIKfyve | ||
| Type: protein-coding gene | Cytoband: 16q22.1-q22.2 | ||
| Entrez ID: 55697 | HGNC ID: HGNC:25507 | Ensembl Gene: ENSG00000103043 | OMIM ID: 604632 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of VAC14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VAC14 | 55697 | 218169_at | 0.6526 | 0.0206 | |
| GSE20347 | VAC14 | 55697 | 218169_at | 0.3954 | 0.0000 | |
| GSE23400 | VAC14 | 55697 | 218169_at | 0.2759 | 0.0000 | |
| GSE26886 | VAC14 | 55697 | 218169_at | 0.6454 | 0.0003 | |
| GSE29001 | VAC14 | 55697 | 218169_at | -0.0938 | 0.6915 | |
| GSE38129 | VAC14 | 55697 | 218169_at | 0.3793 | 0.0001 | |
| GSE45670 | VAC14 | 55697 | 218169_at | 0.3156 | 0.0005 | |
| GSE53622 | VAC14 | 55697 | 30569 | 0.7084 | 0.0000 | |
| GSE53624 | VAC14 | 55697 | 89234 | 0.6956 | 0.0000 | |
| GSE63941 | VAC14 | 55697 | 218169_at | 1.2008 | 0.0135 | |
| GSE77861 | VAC14 | 55697 | 218169_at | 0.5052 | 0.0242 | |
| GSE97050 | VAC14 | 55697 | A_23_P77593 | 0.8604 | 0.0599 | |
| SRP007169 | VAC14 | 55697 | RNAseq | 0.3146 | 0.3179 | |
| SRP008496 | VAC14 | 55697 | RNAseq | 0.3279 | 0.1958 | |
| SRP064894 | VAC14 | 55697 | RNAseq | 0.6482 | 0.0001 | |
| SRP133303 | VAC14 | 55697 | RNAseq | 0.6747 | 0.0000 | |
| SRP159526 | VAC14 | 55697 | RNAseq | 0.5895 | 0.0063 | |
| SRP193095 | VAC14 | 55697 | RNAseq | 0.7835 | 0.0000 | |
| SRP219564 | VAC14 | 55697 | RNAseq | 0.3655 | 0.2565 | |
| TCGA | VAC14 | 55697 | RNAseq | 0.0814 | 0.0744 |
Upregulated datasets: 1; Downregulated datasets: 0.
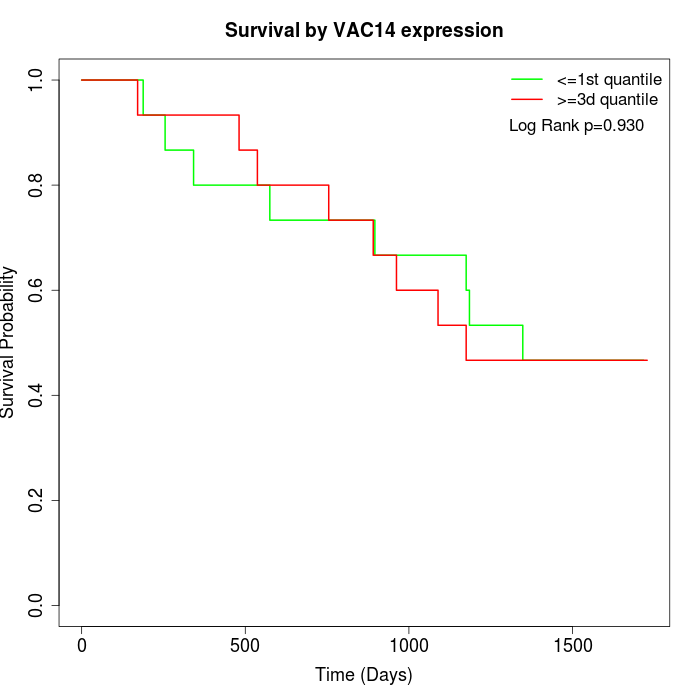
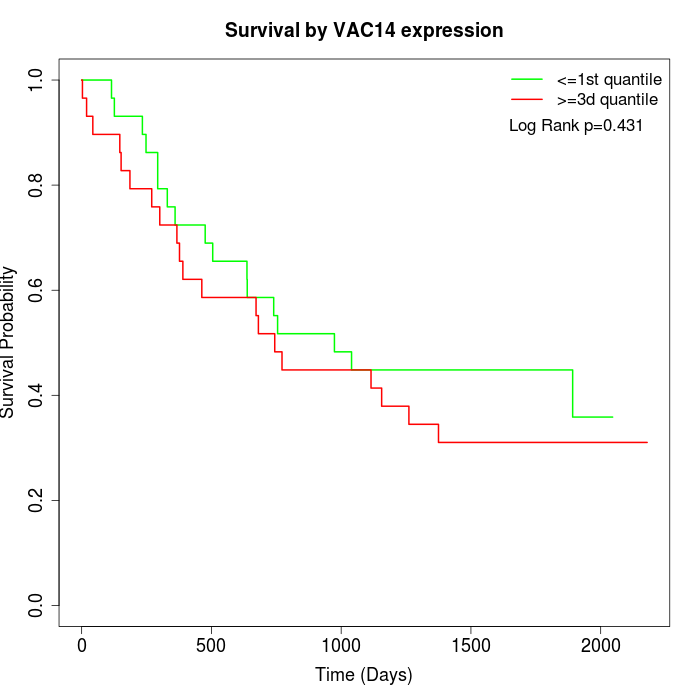
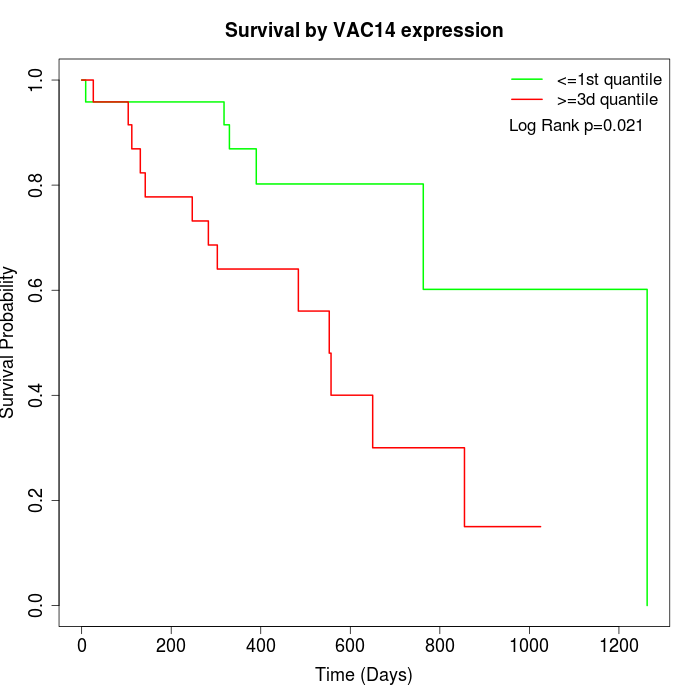
Survival by VAC14 expression:
Note: Click image to view full size file.
Copy number change of VAC14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VAC14 | 55697 | 6 | 1 | 23 | |
| GSE20123 | VAC14 | 55697 | 6 | 1 | 23 | |
| GSE43470 | VAC14 | 55697 | 2 | 8 | 33 | |
| GSE46452 | VAC14 | 55697 | 38 | 1 | 20 | |
| GSE47630 | VAC14 | 55697 | 11 | 8 | 21 | |
| GSE54993 | VAC14 | 55697 | 2 | 4 | 64 | |
| GSE54994 | VAC14 | 55697 | 8 | 10 | 35 | |
| GSE60625 | VAC14 | 55697 | 4 | 0 | 7 | |
| GSE74703 | VAC14 | 55697 | 2 | 5 | 29 | |
| GSE74704 | VAC14 | 55697 | 3 | 0 | 17 | |
| TCGA | VAC14 | 55697 | 27 | 12 | 57 |
Total number of gains: 109; Total number of losses: 50; Total Number of normals: 329.
Somatic mutations of VAC14:
Generating mutation plots.
Highly correlated genes for VAC14:
Showing top 20/1614 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VAC14 | ZNF358 | 0.770472 | 3 | 0 | 3 |
| VAC14 | TRADD | 0.731865 | 4 | 0 | 4 |
| VAC14 | PLEKHG2 | 0.729879 | 3 | 0 | 3 |
| VAC14 | RHOBTB2 | 0.728951 | 3 | 0 | 3 |
| VAC14 | CDCA2 | 0.719614 | 4 | 0 | 4 |
| VAC14 | RILPL1 | 0.71945 | 3 | 0 | 3 |
| VAC14 | SPNS1 | 0.717928 | 4 | 0 | 4 |
| VAC14 | SMYD5 | 0.714601 | 10 | 0 | 10 |
| VAC14 | ZNF628 | 0.7113 | 5 | 0 | 4 |
| VAC14 | EBNA1BP2 | 0.709822 | 3 | 0 | 3 |
| VAC14 | BANF1 | 0.707799 | 12 | 0 | 12 |
| VAC14 | TMEM170A | 0.706459 | 3 | 0 | 3 |
| VAC14 | PSKH1 | 0.704382 | 4 | 0 | 4 |
| VAC14 | LEMD2 | 0.703804 | 4 | 0 | 4 |
| VAC14 | TANGO2 | 0.702644 | 5 | 0 | 4 |
| VAC14 | NSD1 | 0.701649 | 3 | 0 | 3 |
| VAC14 | SIRPG | 0.701009 | 3 | 0 | 3 |
| VAC14 | SPIRE1 | 0.700102 | 3 | 0 | 3 |
| VAC14 | MSTO1 | 0.699161 | 5 | 0 | 5 |
| VAC14 | PROX1 | 0.698303 | 4 | 0 | 4 |
For details and further investigation, click here