| Full name: vinculin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q22.2 | ||
| Entrez ID: 7414 | HGNC ID: HGNC:12665 | Ensembl Gene: ENSG00000035403 | OMIM ID: 193065 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
VCL involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04510 | Focal adhesion | |
| hsa04520 | Adherens junction | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05100 | Bacterial invasion of epithelial cells |
Expression of VCL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VCL | 7414 | 200931_s_at | -0.5078 | 0.4861 | |
| GSE20347 | VCL | 7414 | 200931_s_at | 0.0895 | 0.7119 | |
| GSE23400 | VCL | 7414 | 200931_s_at | -0.1002 | 0.4067 | |
| GSE26886 | VCL | 7414 | 200931_s_at | 0.0631 | 0.7201 | |
| GSE29001 | VCL | 7414 | 200931_s_at | -0.0253 | 0.9133 | |
| GSE38129 | VCL | 7414 | 200931_s_at | -0.2109 | 0.4070 | |
| GSE45670 | VCL | 7414 | 200931_s_at | -0.6343 | 0.0005 | |
| GSE53622 | VCL | 7414 | 107362 | -0.1376 | 0.1560 | |
| GSE53624 | VCL | 7414 | 107362 | 0.0072 | 0.9205 | |
| GSE63941 | VCL | 7414 | 200931_s_at | -1.9041 | 0.0884 | |
| GSE77861 | VCL | 7414 | 200931_s_at | 0.2540 | 0.4326 | |
| GSE97050 | VCL | 7414 | A_24_P47182 | -0.4337 | 0.3058 | |
| SRP007169 | VCL | 7414 | RNAseq | 0.3098 | 0.3856 | |
| SRP008496 | VCL | 7414 | RNAseq | 0.6386 | 0.0026 | |
| SRP064894 | VCL | 7414 | RNAseq | -0.4632 | 0.0647 | |
| SRP133303 | VCL | 7414 | RNAseq | 0.1642 | 0.4675 | |
| SRP159526 | VCL | 7414 | RNAseq | -0.6801 | 0.0926 | |
| SRP193095 | VCL | 7414 | RNAseq | 0.5150 | 0.0003 | |
| SRP219564 | VCL | 7414 | RNAseq | -0.3493 | 0.5937 | |
| TCGA | VCL | 7414 | RNAseq | -0.1013 | 0.0828 |
Upregulated datasets: 0; Downregulated datasets: 0.
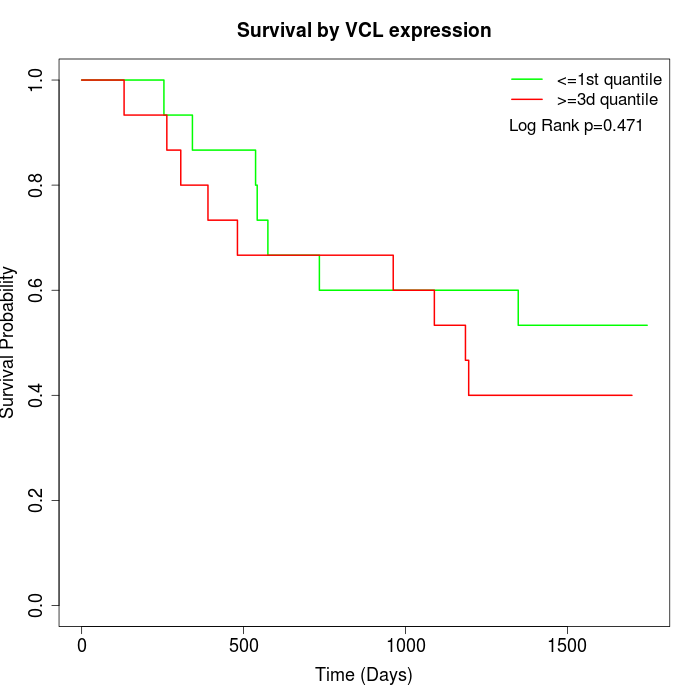
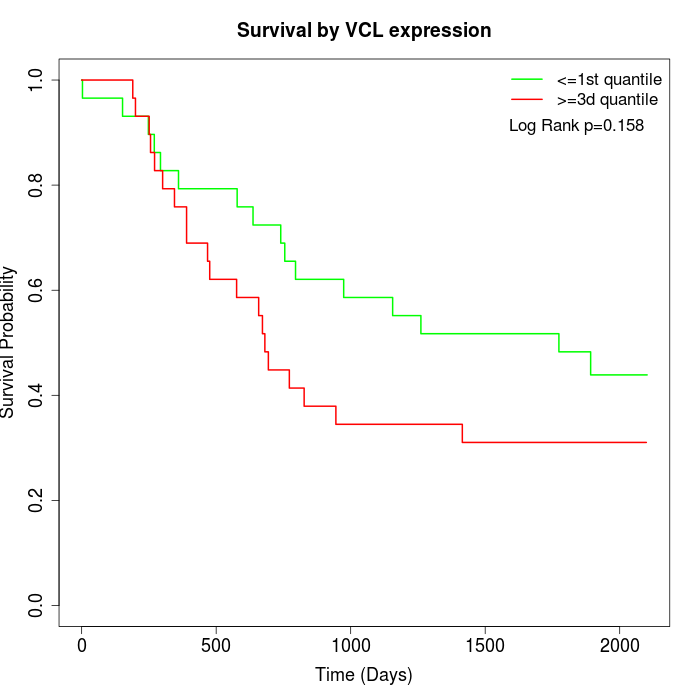
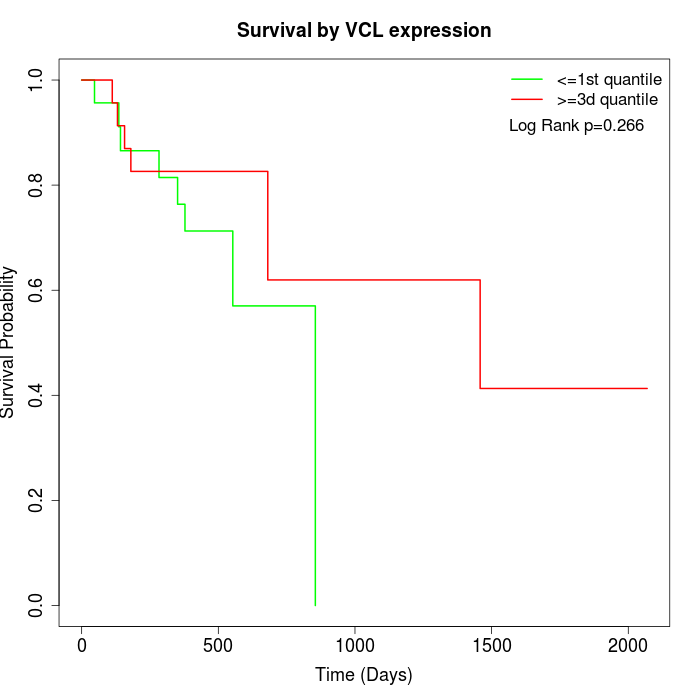
Survival by VCL expression:
Note: Click image to view full size file.
Copy number change of VCL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VCL | 7414 | 3 | 4 | 23 | |
| GSE20123 | VCL | 7414 | 3 | 3 | 24 | |
| GSE43470 | VCL | 7414 | 1 | 8 | 34 | |
| GSE46452 | VCL | 7414 | 0 | 11 | 48 | |
| GSE47630 | VCL | 7414 | 2 | 14 | 24 | |
| GSE54993 | VCL | 7414 | 7 | 0 | 63 | |
| GSE54994 | VCL | 7414 | 2 | 10 | 41 | |
| GSE60625 | VCL | 7414 | 0 | 0 | 11 | |
| GSE74703 | VCL | 7414 | 1 | 5 | 30 | |
| GSE74704 | VCL | 7414 | 1 | 1 | 18 | |
| TCGA | VCL | 7414 | 10 | 23 | 63 |
Total number of gains: 30; Total number of losses: 79; Total Number of normals: 379.
Somatic mutations of VCL:
Generating mutation plots.
Highly correlated genes for VCL:
Showing top 20/861 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VCL | NDUFB2 | 0.778544 | 3 | 0 | 3 |
| VCL | TMOD1 | 0.751357 | 5 | 0 | 4 |
| VCL | FILIP1 | 0.743706 | 5 | 0 | 5 |
| VCL | MRVI1 | 0.738729 | 5 | 0 | 5 |
| VCL | MYADM | 0.738163 | 5 | 0 | 4 |
| VCL | FAM122C | 0.736737 | 3 | 0 | 3 |
| VCL | BVES | 0.735705 | 5 | 0 | 5 |
| VCL | RASSF8 | 0.731669 | 5 | 0 | 5 |
| VCL | TSPAN15 | 0.719739 | 3 | 0 | 3 |
| VCL | CYS1 | 0.717298 | 5 | 0 | 4 |
| VCL | MYOCD | 0.714426 | 5 | 0 | 4 |
| VCL | EPM2A | 0.714337 | 5 | 0 | 5 |
| VCL | FLNA | 0.713956 | 10 | 0 | 9 |
| VCL | CPXM2 | 0.713603 | 4 | 0 | 4 |
| VCL | SGCA | 0.712843 | 6 | 0 | 6 |
| VCL | PARVA | 0.712575 | 9 | 0 | 8 |
| VCL | PALLD | 0.712094 | 11 | 0 | 9 |
| VCL | AMOTL1 | 0.710976 | 4 | 0 | 4 |
| VCL | PPP1R12B | 0.709886 | 6 | 0 | 5 |
| VCL | FERMT2 | 0.709395 | 11 | 0 | 9 |
For details and further investigation, click here