| Full name: protein phosphatase 1 regulatory subunit 12B | Alias Symbol: MGC87886|MGC131980|PP1bp55 | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 4660 | HGNC ID: HGNC:7619 | Ensembl Gene: ENSG00000077157 | OMIM ID: 603768 |
| Drug and gene relationship at DGIdb | |||
PPP1R12B involved pathways:
Expression of PPP1R12B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPP1R12B | 4660 | 201957_at | -1.6910 | 0.1611 | |
| GSE20347 | PPP1R12B | 4660 | 201957_at | -0.5407 | 0.0735 | |
| GSE23400 | PPP1R12B | 4660 | 201957_at | -0.8235 | 0.0000 | |
| GSE26886 | PPP1R12B | 4660 | 201957_at | 0.0953 | 0.8226 | |
| GSE29001 | PPP1R12B | 4660 | 201957_at | -0.7723 | 0.0339 | |
| GSE38129 | PPP1R12B | 4660 | 201957_at | -1.1972 | 0.0146 | |
| GSE45670 | PPP1R12B | 4660 | 201957_at | -2.5633 | 0.0000 | |
| GSE53622 | PPP1R12B | 4660 | 13339 | -2.5014 | 0.0000 | |
| GSE53624 | PPP1R12B | 4660 | 13339 | -2.2964 | 0.0000 | |
| GSE63941 | PPP1R12B | 4660 | 201957_at | -0.9661 | 0.0680 | |
| GSE77861 | PPP1R12B | 4660 | 201957_at | 0.0426 | 0.8739 | |
| GSE97050 | PPP1R12B | 4660 | A_33_P3257609 | -1.9813 | 0.0844 | |
| SRP007169 | PPP1R12B | 4660 | RNAseq | 0.3293 | 0.5120 | |
| SRP008496 | PPP1R12B | 4660 | RNAseq | 0.8116 | 0.0412 | |
| SRP064894 | PPP1R12B | 4660 | RNAseq | -0.8105 | 0.0471 | |
| SRP133303 | PPP1R12B | 4660 | RNAseq | -1.7577 | 0.0000 | |
| SRP159526 | PPP1R12B | 4660 | RNAseq | -1.5473 | 0.0104 | |
| SRP193095 | PPP1R12B | 4660 | RNAseq | -0.2716 | 0.0863 | |
| SRP219564 | PPP1R12B | 4660 | RNAseq | -0.7755 | 0.4356 | |
| TCGA | PPP1R12B | 4660 | RNAseq | -0.6477 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 6.
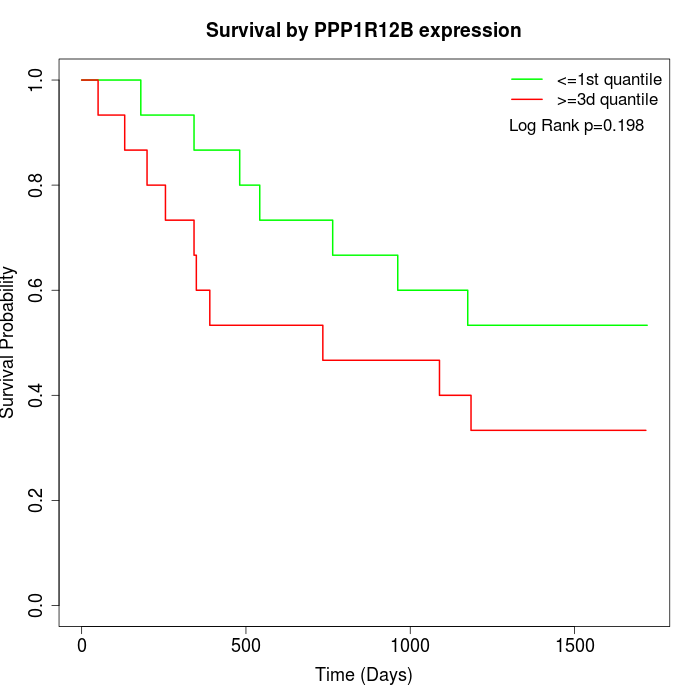
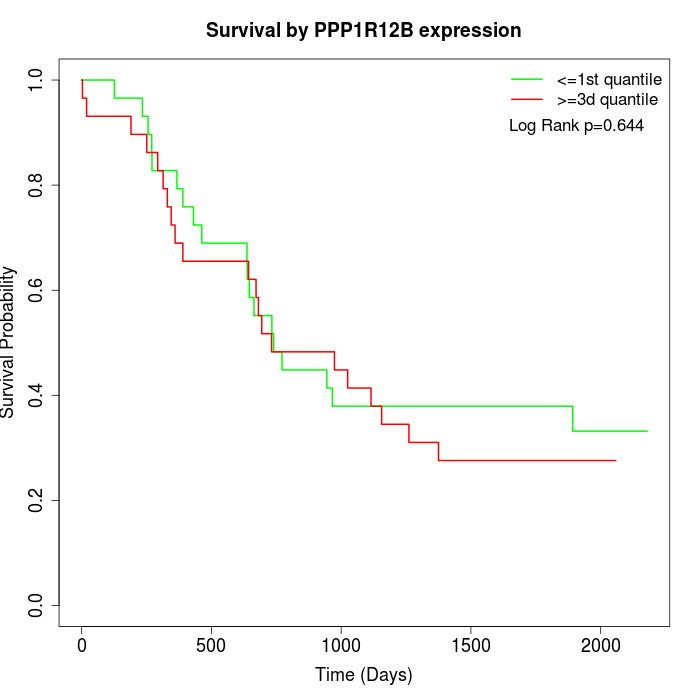
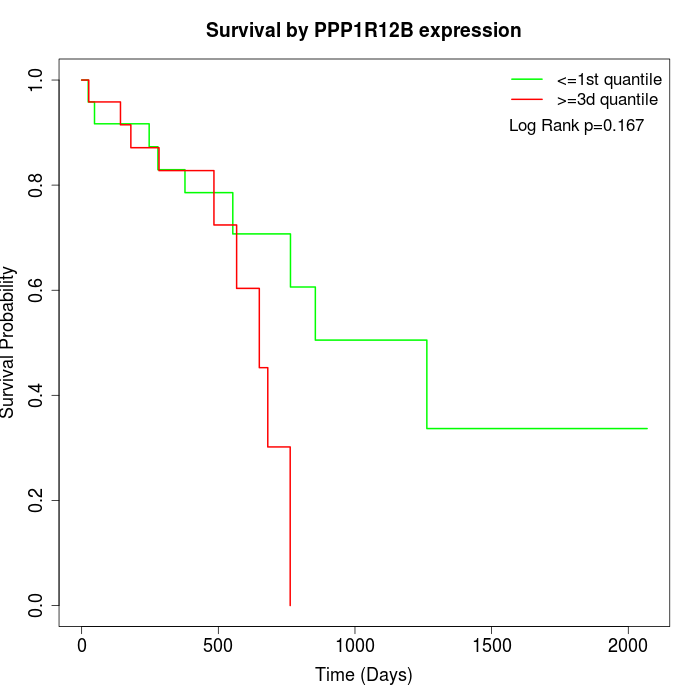
Survival by PPP1R12B expression:
Note: Click image to view full size file.
Copy number change of PPP1R12B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPP1R12B | 4660 | 11 | 0 | 19 | |
| GSE20123 | PPP1R12B | 4660 | 11 | 0 | 19 | |
| GSE43470 | PPP1R12B | 4660 | 6 | 0 | 37 | |
| GSE46452 | PPP1R12B | 4660 | 3 | 1 | 55 | |
| GSE47630 | PPP1R12B | 4660 | 14 | 0 | 26 | |
| GSE54993 | PPP1R12B | 4660 | 0 | 6 | 64 | |
| GSE54994 | PPP1R12B | 4660 | 15 | 0 | 38 | |
| GSE60625 | PPP1R12B | 4660 | 0 | 0 | 11 | |
| GSE74703 | PPP1R12B | 4660 | 6 | 0 | 30 | |
| GSE74704 | PPP1R12B | 4660 | 5 | 0 | 15 | |
| TCGA | PPP1R12B | 4660 | 43 | 4 | 49 |
Total number of gains: 114; Total number of losses: 11; Total Number of normals: 363.
Somatic mutations of PPP1R12B:
Generating mutation plots.
Highly correlated genes for PPP1R12B:
Showing top 20/1263 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPP1R12B | ATP1A2 | 0.896983 | 9 | 0 | 9 |
| PPP1R12B | DAAM2 | 0.881559 | 8 | 0 | 8 |
| PPP1R12B | XKR4 | 0.869867 | 5 | 0 | 5 |
| PPP1R12B | LMOD1 | 0.861628 | 12 | 0 | 11 |
| PPP1R12B | SORCS1 | 0.860777 | 4 | 0 | 4 |
| PPP1R12B | SYNM | 0.841771 | 11 | 0 | 11 |
| PPP1R12B | DES | 0.840345 | 11 | 0 | 10 |
| PPP1R12B | LPP | 0.839435 | 8 | 0 | 7 |
| PPP1R12B | CYP21A2 | 0.837833 | 3 | 0 | 3 |
| PPP1R12B | RAB9B | 0.834807 | 6 | 0 | 6 |
| PPP1R12B | CASQ2 | 0.831402 | 11 | 0 | 11 |
| PPP1R12B | ACTBL2 | 0.828357 | 3 | 0 | 3 |
| PPP1R12B | BCHE | 0.825488 | 8 | 0 | 8 |
| PPP1R12B | NTN1 | 0.82506 | 5 | 0 | 5 |
| PPP1R12B | ITGA7 | 0.824363 | 10 | 0 | 10 |
| PPP1R12B | CACNB2 | 0.824063 | 8 | 0 | 8 |
| PPP1R12B | HSPB7 | 0.823623 | 9 | 0 | 8 |
| PPP1R12B | CDH19 | 0.82265 | 8 | 0 | 8 |
| PPP1R12B | CNN1 | 0.822461 | 10 | 0 | 9 |
| PPP1R12B | CHRDL1 | 0.822084 | 9 | 0 | 9 |
For details and further investigation, click here