| Full name: vitrin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2p22.2 | ||
| Entrez ID: 5212 | HGNC ID: HGNC:12697 | Ensembl Gene: ENSG00000205221 | OMIM ID: 617693 |
| Drug and gene relationship at DGIdb | |||
Expression of VIT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VIT | 5212 | 227899_at | -2.1389 | 0.0124 | |
| GSE26886 | VIT | 5212 | 227899_at | -0.8761 | 0.0060 | |
| GSE45670 | VIT | 5212 | 227899_at | -2.1896 | 0.0000 | |
| GSE53622 | VIT | 5212 | 84032 | -1.8396 | 0.0000 | |
| GSE53624 | VIT | 5212 | 84032 | -1.6549 | 0.0000 | |
| GSE63941 | VIT | 5212 | 227899_at | -0.0754 | 0.8878 | |
| GSE77861 | VIT | 5212 | 227899_at | -0.5414 | 0.0038 | |
| GSE97050 | VIT | 5212 | A_23_P56578 | -1.2498 | 0.1210 | |
| SRP007169 | VIT | 5212 | RNAseq | -2.7927 | 0.0000 | |
| SRP064894 | VIT | 5212 | RNAseq | -2.8177 | 0.0000 | |
| SRP133303 | VIT | 5212 | RNAseq | -3.4205 | 0.0000 | |
| SRP193095 | VIT | 5212 | RNAseq | -3.6813 | 0.0000 | |
| SRP219564 | VIT | 5212 | RNAseq | -1.3455 | 0.2466 | |
| TCGA | VIT | 5212 | RNAseq | -1.1875 | 0.0532 |
Upregulated datasets: 0; Downregulated datasets: 8.
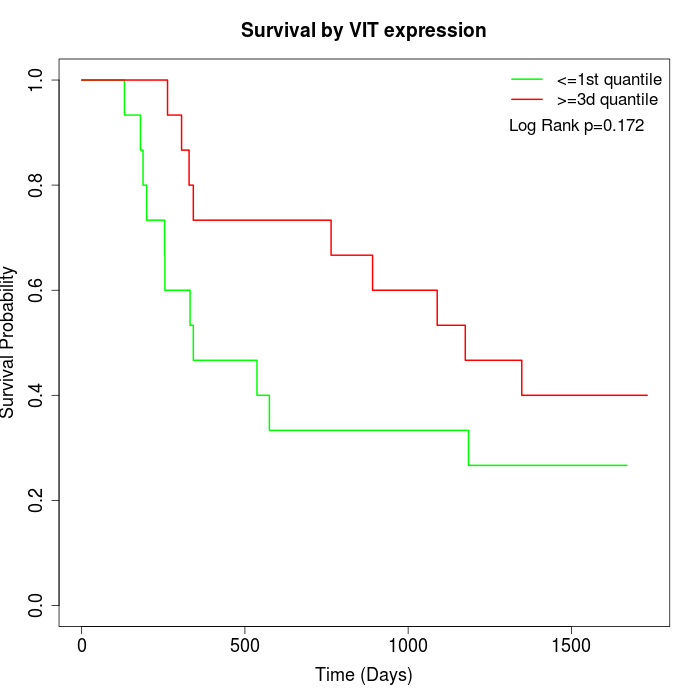
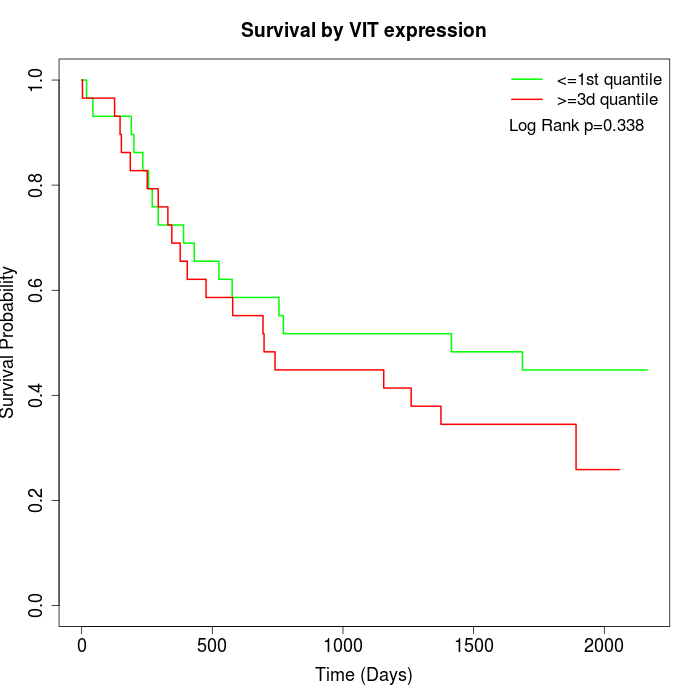
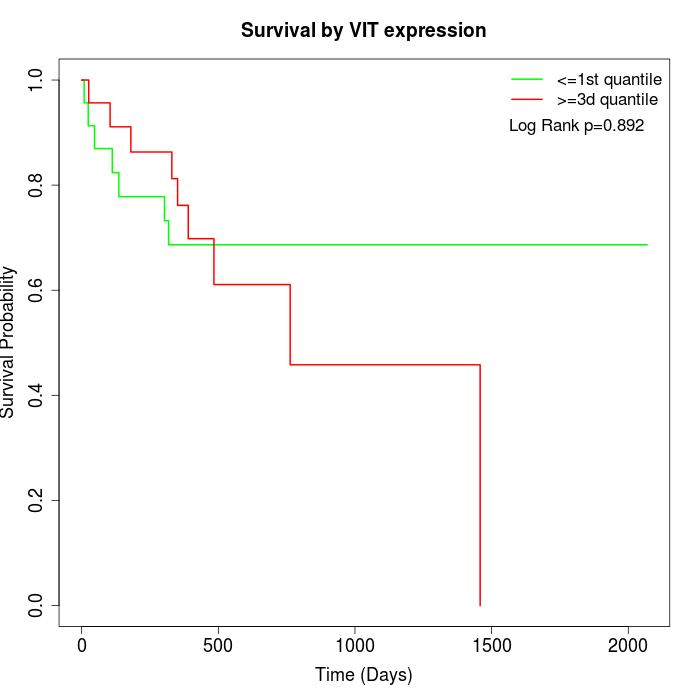
Survival by VIT expression:
Note: Click image to view full size file.
Copy number change of VIT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VIT | 5212 | 10 | 1 | 19 | |
| GSE20123 | VIT | 5212 | 8 | 1 | 21 | |
| GSE43470 | VIT | 5212 | 3 | 0 | 40 | |
| GSE46452 | VIT | 5212 | 1 | 4 | 54 | |
| GSE47630 | VIT | 5212 | 7 | 0 | 33 | |
| GSE54993 | VIT | 5212 | 0 | 5 | 65 | |
| GSE54994 | VIT | 5212 | 10 | 0 | 43 | |
| GSE60625 | VIT | 5212 | 0 | 3 | 8 | |
| GSE74703 | VIT | 5212 | 3 | 0 | 33 | |
| GSE74704 | VIT | 5212 | 8 | 1 | 11 | |
| TCGA | VIT | 5212 | 35 | 2 | 59 |
Total number of gains: 85; Total number of losses: 17; Total Number of normals: 386.
Somatic mutations of VIT:
Generating mutation plots.
Highly correlated genes for VIT:
Showing top 20/1183 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VIT | CFD | 0.804239 | 5 | 0 | 5 |
| VIT | CHRDL1 | 0.801179 | 4 | 0 | 4 |
| VIT | TMOD1 | 0.786675 | 3 | 0 | 3 |
| VIT | SCN7A | 0.784935 | 5 | 0 | 5 |
| VIT | FXYD1 | 0.771382 | 4 | 0 | 3 |
| VIT | FOXP1 | 0.762815 | 4 | 0 | 4 |
| VIT | PHC1 | 0.762182 | 5 | 0 | 5 |
| VIT | RERGL | 0.75815 | 5 | 0 | 5 |
| VIT | ANGPTL1 | 0.757924 | 5 | 0 | 5 |
| VIT | ACOX2 | 0.75685 | 6 | 0 | 6 |
| VIT | NEGR1 | 0.754729 | 5 | 0 | 5 |
| VIT | DIP2C | 0.754688 | 3 | 0 | 3 |
| VIT | C7 | 0.752617 | 5 | 0 | 5 |
| VIT | BOC | 0.752532 | 6 | 0 | 6 |
| VIT | RAB9B | 0.751641 | 5 | 0 | 5 |
| VIT | SSBP2 | 0.750106 | 6 | 0 | 6 |
| VIT | SLC27A6 | 0.749849 | 7 | 0 | 7 |
| VIT | RCAN2 | 0.748464 | 5 | 0 | 5 |
| VIT | PCP4 | 0.746484 | 5 | 0 | 5 |
| VIT | GPIHBP1 | 0.745322 | 3 | 0 | 3 |
For details and further investigation, click here