| Full name: complement factor D | Alias Symbol: ADN | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 1675 | HGNC ID: HGNC:2771 | Ensembl Gene: ENSG00000197766 | OMIM ID: 134350 |
| Drug and gene relationship at DGIdb | |||
CFD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04610 | Complement and coagulation cascades | |
| hsa05150 | Staphylococcus aureus infection |
Expression of CFD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFD | 1675 | 205382_s_at | -2.9039 | 0.0265 | |
| GSE20347 | CFD | 1675 | 205382_s_at | -1.0932 | 0.0001 | |
| GSE23400 | CFD | 1675 | 205382_s_at | -2.2629 | 0.0000 | |
| GSE26886 | CFD | 1675 | 205382_s_at | -0.2694 | 0.4320 | |
| GSE29001 | CFD | 1675 | 205382_s_at | -1.9858 | 0.0014 | |
| GSE38129 | CFD | 1675 | 205382_s_at | -2.3172 | 0.0000 | |
| GSE45670 | CFD | 1675 | 205382_s_at | -3.5517 | 0.0000 | |
| GSE53622 | CFD | 1675 | 16314 | -2.5883 | 0.0000 | |
| GSE53624 | CFD | 1675 | 16314 | -2.1122 | 0.0000 | |
| GSE63941 | CFD | 1675 | 205382_s_at | -2.2570 | 0.0248 | |
| GSE77861 | CFD | 1675 | 205382_s_at | -0.2205 | 0.6002 | |
| GSE97050 | CFD | 1675 | A_23_P119562 | -1.2739 | 0.0555 | |
| SRP007169 | CFD | 1675 | RNAseq | -2.1043 | 0.0003 | |
| SRP008496 | CFD | 1675 | RNAseq | -2.3608 | 0.0000 | |
| SRP064894 | CFD | 1675 | RNAseq | -2.0814 | 0.0028 | |
| SRP133303 | CFD | 1675 | RNAseq | -2.8939 | 0.0000 | |
| SRP159526 | CFD | 1675 | RNAseq | -2.6864 | 0.0001 | |
| SRP193095 | CFD | 1675 | RNAseq | -0.8832 | 0.0001 | |
| SRP219564 | CFD | 1675 | RNAseq | -0.8556 | 0.3734 | |
| TCGA | CFD | 1675 | RNAseq | -0.9785 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 14.
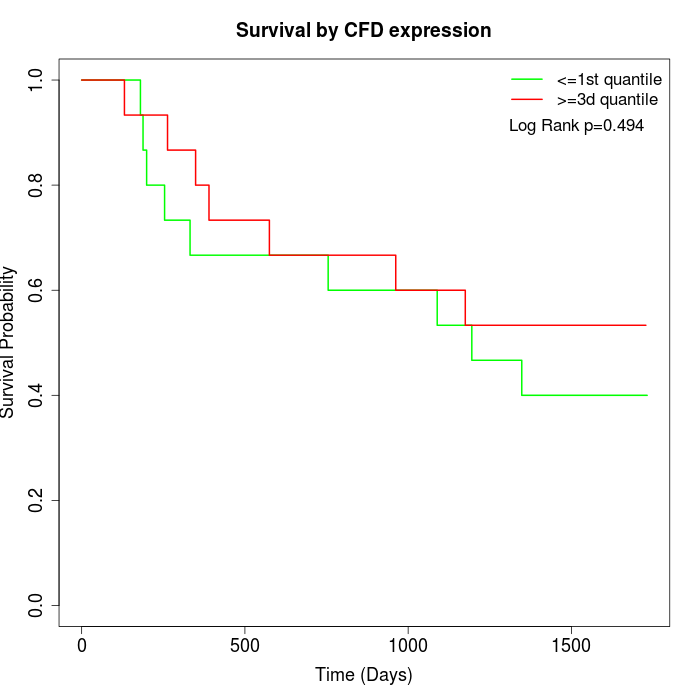
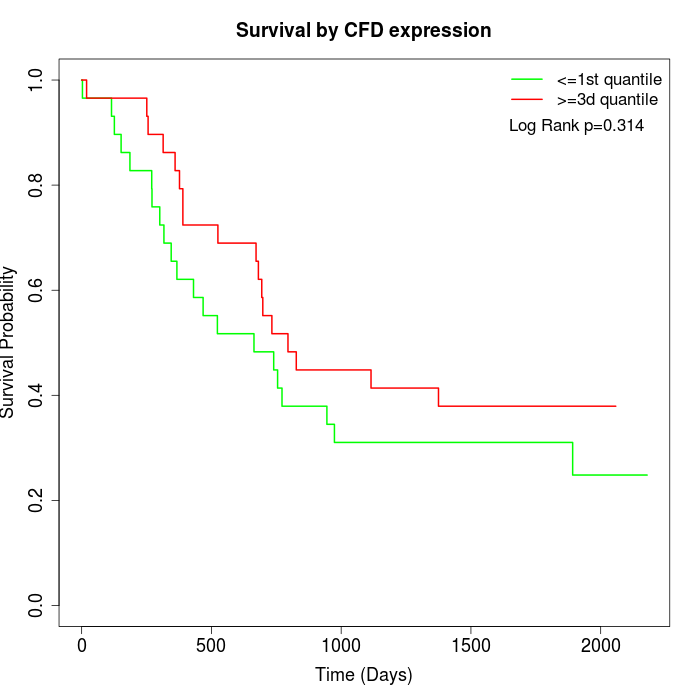
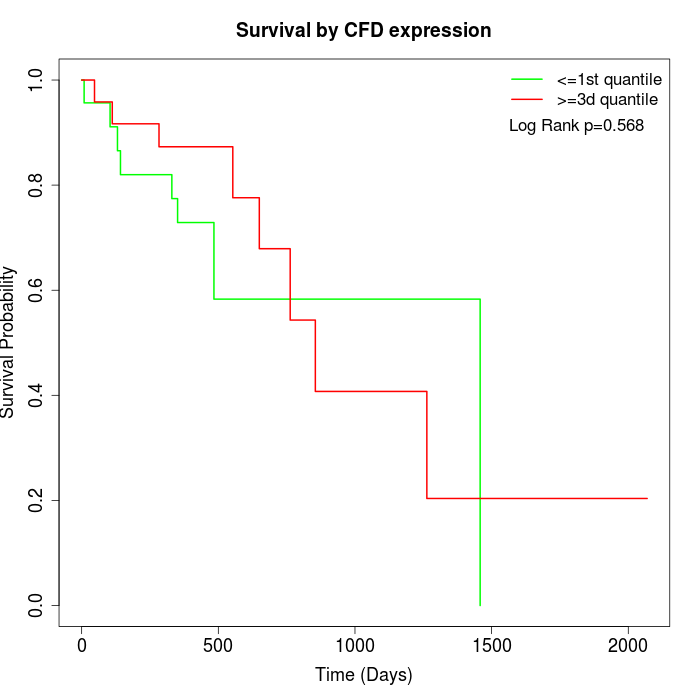
Survival by CFD expression:
Note: Click image to view full size file.
Copy number change of CFD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFD | 1675 | 4 | 6 | 20 | |
| GSE20123 | CFD | 1675 | 4 | 5 | 21 | |
| GSE43470 | CFD | 1675 | 1 | 9 | 33 | |
| GSE46452 | CFD | 1675 | 47 | 1 | 11 | |
| GSE47630 | CFD | 1675 | 5 | 7 | 28 | |
| GSE54993 | CFD | 1675 | 13 | 4 | 53 | |
| GSE54994 | CFD | 1675 | 8 | 15 | 30 | |
| GSE60625 | CFD | 1675 | 9 | 0 | 2 | |
| GSE74703 | CFD | 1675 | 1 | 7 | 28 | |
| GSE74704 | CFD | 1675 | 3 | 3 | 14 | |
| TCGA | CFD | 1675 | 8 | 25 | 63 |
Total number of gains: 103; Total number of losses: 82; Total Number of normals: 303.
Somatic mutations of CFD:
Generating mutation plots.
Highly correlated genes for CFD:
Showing top 20/1567 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFD | CLEC3B | 0.930322 | 3 | 0 | 3 |
| CFD | CYP21A2 | 0.909058 | 3 | 0 | 3 |
| CFD | MAMDC2 | 0.865798 | 5 | 0 | 5 |
| CFD | SORCS1 | 0.849581 | 4 | 0 | 4 |
| CFD | CCL14 | 0.842496 | 3 | 0 | 3 |
| CFD | ANGPTL1 | 0.83488 | 6 | 0 | 6 |
| CFD | XKR4 | 0.827359 | 5 | 0 | 5 |
| CFD | RAB9B | 0.807406 | 5 | 0 | 5 |
| CFD | VIT | 0.804239 | 5 | 0 | 5 |
| CFD | RGS7BP | 0.797037 | 5 | 0 | 5 |
| CFD | ATP1A2 | 0.796819 | 9 | 0 | 9 |
| CFD | SLMAP | 0.79203 | 5 | 0 | 5 |
| CFD | C7 | 0.788018 | 9 | 0 | 8 |
| CFD | ABCA8 | 0.786643 | 10 | 0 | 10 |
| CFD | CHRDL1 | 0.783255 | 8 | 0 | 8 |
| CFD | MYRIP | 0.781773 | 9 | 0 | 9 |
| CFD | GSN | 0.774473 | 9 | 0 | 9 |
| CFD | F10 | 0.773852 | 10 | 0 | 9 |
| CFD | CASQ2 | 0.77368 | 9 | 0 | 9 |
| CFD | FAM13C | 0.772586 | 4 | 0 | 4 |
For details and further investigation, click here