| Full name: YES proto-oncogene 1, Src family tyrosine kinase | Alias Symbol: Yes|c-yes|HsT441 | ||
| Type: protein-coding gene | Cytoband: 18p11.32 | ||
| Entrez ID: 7525 | HGNC ID: HGNC:12841 | Ensembl Gene: ENSG00000176105 | OMIM ID: 164880 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
YES1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04520 | Adherens junction |
Expression of YES1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | YES1 | 7525 | 202933_s_at | 0.3521 | 0.6916 | |
| GSE20347 | YES1 | 7525 | 202933_s_at | 0.2282 | 0.1028 | |
| GSE23400 | YES1 | 7525 | 202932_at | 0.3211 | 0.0015 | |
| GSE26886 | YES1 | 7525 | 202933_s_at | 0.0815 | 0.7073 | |
| GSE29001 | YES1 | 7525 | 202933_s_at | 0.2448 | 0.1973 | |
| GSE38129 | YES1 | 7525 | 202933_s_at | 0.2829 | 0.0816 | |
| GSE45670 | YES1 | 7525 | 202933_s_at | 0.2520 | 0.0344 | |
| GSE53622 | YES1 | 7525 | 32477 | 0.2215 | 0.0129 | |
| GSE53624 | YES1 | 7525 | 32477 | 0.1733 | 0.0694 | |
| GSE63941 | YES1 | 7525 | 202933_s_at | 0.4463 | 0.3949 | |
| GSE77861 | YES1 | 7525 | 202933_s_at | 0.2863 | 0.0791 | |
| GSE97050 | YES1 | 7525 | A_24_P48403 | 0.3174 | 0.3225 | |
| SRP007169 | YES1 | 7525 | RNAseq | 0.0240 | 0.9453 | |
| SRP008496 | YES1 | 7525 | RNAseq | -0.1010 | 0.6530 | |
| SRP064894 | YES1 | 7525 | RNAseq | 0.2343 | 0.4435 | |
| SRP133303 | YES1 | 7525 | RNAseq | 0.7393 | 0.0003 | |
| SRP159526 | YES1 | 7525 | RNAseq | 0.2656 | 0.3776 | |
| SRP193095 | YES1 | 7525 | RNAseq | 0.0837 | 0.5905 | |
| SRP219564 | YES1 | 7525 | RNAseq | -0.3964 | 0.1777 | |
| TCGA | YES1 | 7525 | RNAseq | 0.0841 | 0.1378 |
Upregulated datasets: 0; Downregulated datasets: 0.
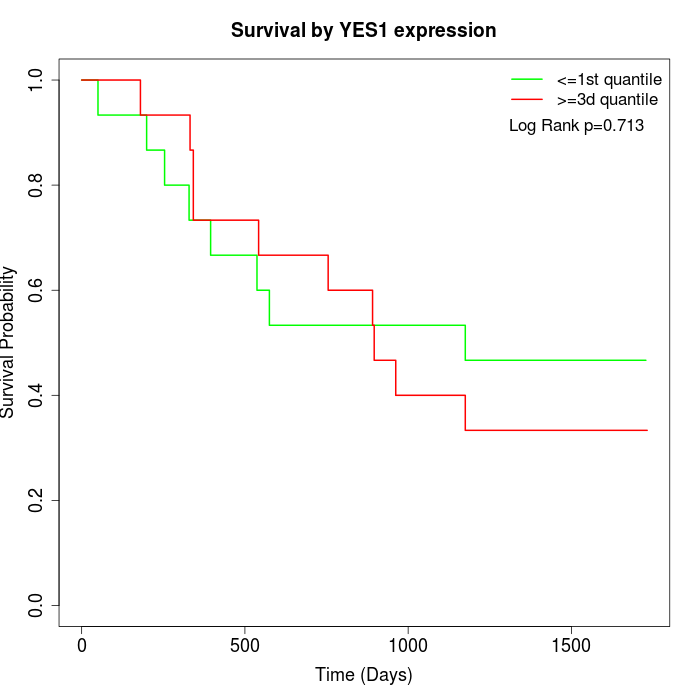
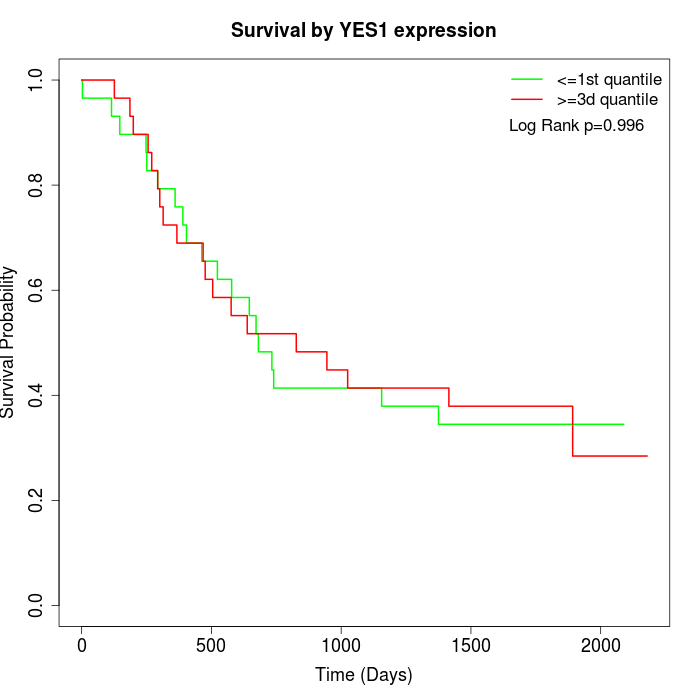
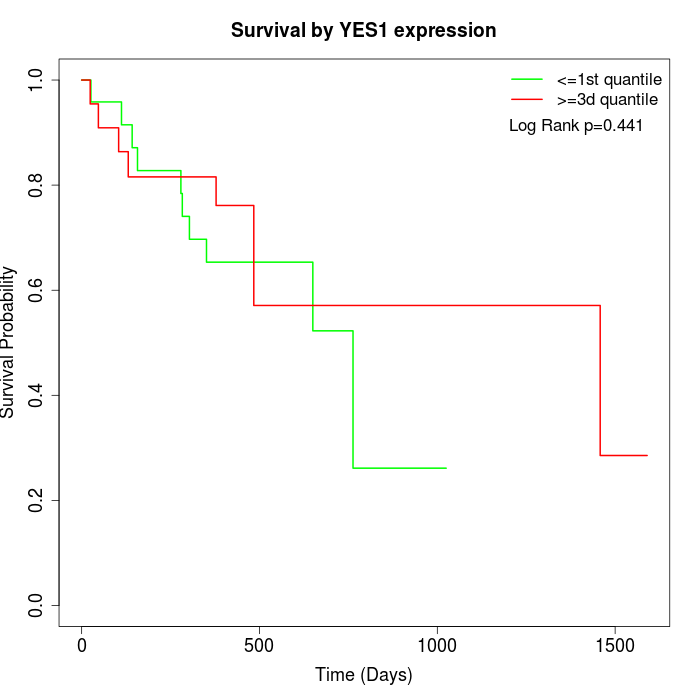
Survival by YES1 expression:
Note: Click image to view full size file.
Copy number change of YES1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | YES1 | 7525 | 6 | 3 | 21 | |
| GSE20123 | YES1 | 7525 | 6 | 3 | 21 | |
| GSE43470 | YES1 | 7525 | 2 | 4 | 37 | |
| GSE46452 | YES1 | 7525 | 4 | 20 | 35 | |
| GSE47630 | YES1 | 7525 | 7 | 18 | 15 | |
| GSE54993 | YES1 | 7525 | 6 | 3 | 61 | |
| GSE54994 | YES1 | 7525 | 11 | 8 | 34 | |
| GSE60625 | YES1 | 7525 | 0 | 4 | 7 | |
| GSE74703 | YES1 | 7525 | 2 | 2 | 32 | |
| GSE74704 | YES1 | 7525 | 4 | 2 | 14 | |
| TCGA | YES1 | 7525 | 35 | 18 | 43 |
Total number of gains: 83; Total number of losses: 85; Total Number of normals: 320.
Somatic mutations of YES1:
Generating mutation plots.
Highly correlated genes for YES1:
Showing top 20/394 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| YES1 | MRPL52 | 0.752696 | 4 | 0 | 4 |
| YES1 | MIPOL1 | 0.732788 | 3 | 0 | 3 |
| YES1 | EAF1 | 0.730157 | 3 | 0 | 3 |
| YES1 | ZNF808 | 0.727277 | 3 | 0 | 3 |
| YES1 | TPCN2 | 0.727102 | 3 | 0 | 3 |
| YES1 | ANAPC13 | 0.726384 | 3 | 0 | 3 |
| YES1 | PPP6R3 | 0.722034 | 4 | 0 | 4 |
| YES1 | PPIL4 | 0.709526 | 3 | 0 | 3 |
| YES1 | IWS1 | 0.70269 | 3 | 0 | 3 |
| YES1 | RASA2 | 0.693257 | 3 | 0 | 3 |
| YES1 | CCDC138 | 0.682163 | 3 | 0 | 3 |
| YES1 | PWP1 | 0.679447 | 3 | 0 | 3 |
| YES1 | PIBF1 | 0.678662 | 4 | 0 | 3 |
| YES1 | SRD5A3 | 0.675599 | 3 | 0 | 3 |
| YES1 | CEP70 | 0.674054 | 3 | 0 | 3 |
| YES1 | GNL3L | 0.669563 | 5 | 0 | 3 |
| YES1 | OPA1 | 0.669177 | 3 | 0 | 3 |
| YES1 | SPPL2A | 0.661772 | 3 | 0 | 3 |
| YES1 | DMTF1 | 0.657454 | 3 | 0 | 3 |
| YES1 | SLC2A9 | 0.651537 | 5 | 0 | 4 |
For details and further investigation, click here