| Full name: OPA1 mitochondrial dynamin like GTPase | Alias Symbol: NTG|KIAA0567|FLJ12460|NPG|MGM1 | ||
| Type: protein-coding gene | Cytoband: 3q29 | ||
| Entrez ID: 4976 | HGNC ID: HGNC:8140 | Ensembl Gene: ENSG00000198836 | OMIM ID: 605290 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of OPA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | OPA1 | 4976 | 212213_x_at | 0.3957 | 0.2968 | |
| GSE20347 | OPA1 | 4976 | 212213_x_at | 0.4374 | 0.0028 | |
| GSE23400 | OPA1 | 4976 | 212213_x_at | 0.3336 | 0.0000 | |
| GSE26886 | OPA1 | 4976 | 212213_x_at | 0.0976 | 0.6355 | |
| GSE29001 | OPA1 | 4976 | 212213_x_at | 0.5587 | 0.0100 | |
| GSE38129 | OPA1 | 4976 | 212213_x_at | 0.2921 | 0.0152 | |
| GSE45670 | OPA1 | 4976 | 212213_x_at | 0.0652 | 0.7790 | |
| GSE53622 | OPA1 | 4976 | 51497 | 0.4917 | 0.0000 | |
| GSE53624 | OPA1 | 4976 | 48178 | 0.3556 | 0.0000 | |
| GSE63941 | OPA1 | 4976 | 212213_x_at | 0.1397 | 0.7820 | |
| GSE77861 | OPA1 | 4976 | 212213_x_at | 0.5590 | 0.1177 | |
| GSE97050 | OPA1 | 4976 | A_33_P3346498 | 0.3568 | 0.2433 | |
| SRP007169 | OPA1 | 4976 | RNAseq | 0.8394 | 0.0217 | |
| SRP008496 | OPA1 | 4976 | RNAseq | 1.0803 | 0.0000 | |
| SRP064894 | OPA1 | 4976 | RNAseq | 0.3073 | 0.1543 | |
| SRP133303 | OPA1 | 4976 | RNAseq | 0.6841 | 0.0002 | |
| SRP159526 | OPA1 | 4976 | RNAseq | 0.7434 | 0.0139 | |
| SRP193095 | OPA1 | 4976 | RNAseq | 0.2644 | 0.0222 | |
| SRP219564 | OPA1 | 4976 | RNAseq | 0.1190 | 0.7093 | |
| TCGA | OPA1 | 4976 | RNAseq | 0.0768 | 0.1016 |
Upregulated datasets: 1; Downregulated datasets: 0.
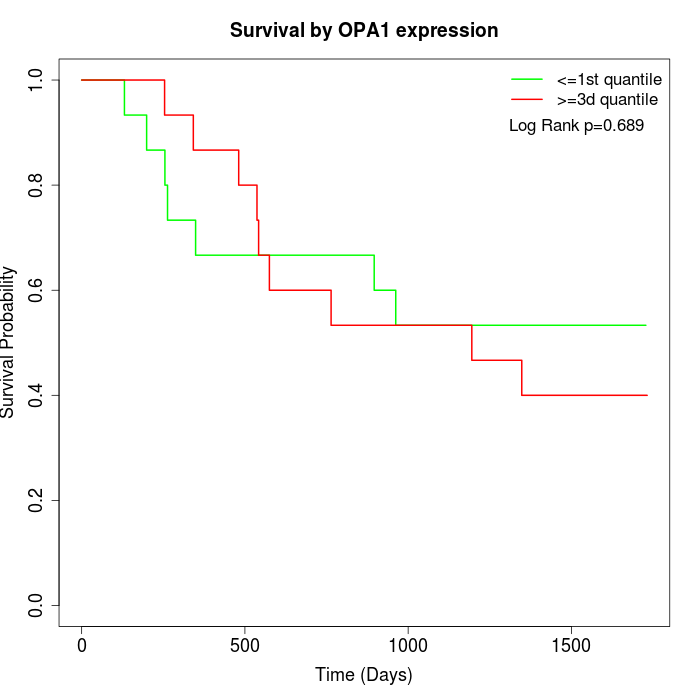
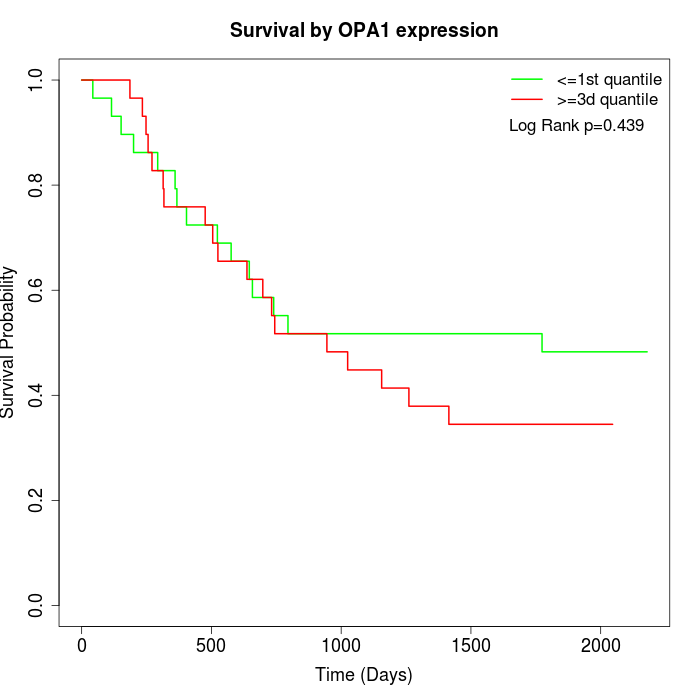
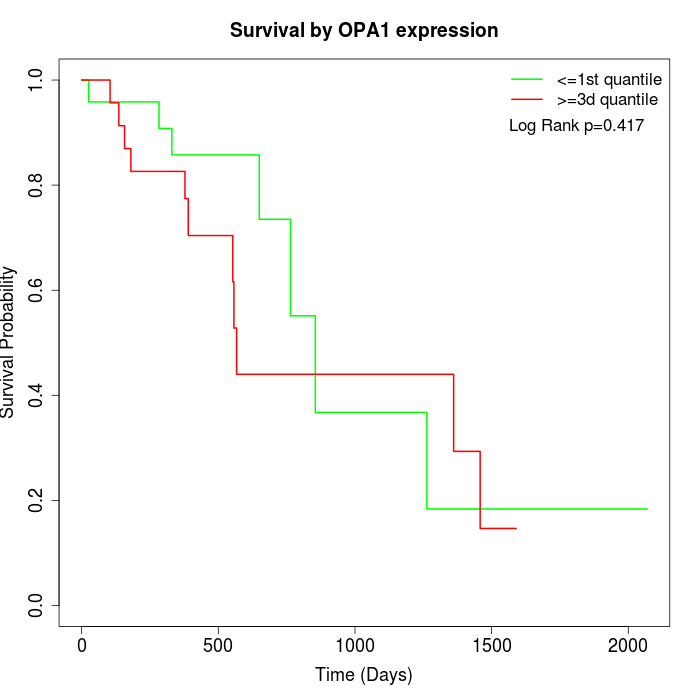
Survival by OPA1 expression:
Note: Click image to view full size file.
Copy number change of OPA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | OPA1 | 4976 | 22 | 0 | 8 | |
| GSE20123 | OPA1 | 4976 | 22 | 0 | 8 | |
| GSE43470 | OPA1 | 4976 | 24 | 0 | 19 | |
| GSE46452 | OPA1 | 4976 | 19 | 1 | 39 | |
| GSE47630 | OPA1 | 4976 | 27 | 2 | 11 | |
| GSE54993 | OPA1 | 4976 | 1 | 20 | 49 | |
| GSE54994 | OPA1 | 4976 | 40 | 0 | 13 | |
| GSE60625 | OPA1 | 4976 | 0 | 6 | 5 | |
| GSE74703 | OPA1 | 4976 | 20 | 0 | 16 | |
| GSE74704 | OPA1 | 4976 | 14 | 0 | 6 | |
| TCGA | OPA1 | 4976 | 77 | 0 | 19 |
Total number of gains: 266; Total number of losses: 29; Total Number of normals: 193.
Somatic mutations of OPA1:
Generating mutation plots.
Highly correlated genes for OPA1:
Showing top 20/875 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| OPA1 | FBXO33 | 0.741342 | 3 | 0 | 3 |
| OPA1 | TAP1 | 0.729454 | 3 | 0 | 3 |
| OPA1 | RAD18 | 0.709513 | 3 | 0 | 3 |
| OPA1 | ZC3HC1 | 0.702051 | 3 | 0 | 3 |
| OPA1 | CXCL13 | 0.693078 | 3 | 0 | 3 |
| OPA1 | EIF3C | 0.692207 | 3 | 0 | 3 |
| OPA1 | LRRC34 | 0.691563 | 3 | 0 | 3 |
| OPA1 | ZSCAN29 | 0.690879 | 3 | 0 | 3 |
| OPA1 | ZGRF1 | 0.681044 | 3 | 0 | 3 |
| OPA1 | PPFIA1 | 0.677885 | 6 | 0 | 5 |
| OPA1 | SRSF10 | 0.674753 | 5 | 0 | 4 |
| OPA1 | YES1 | 0.669177 | 3 | 0 | 3 |
| OPA1 | NCBP2 | 0.664827 | 11 | 0 | 10 |
| OPA1 | CSTF3 | 0.663403 | 5 | 0 | 3 |
| OPA1 | CCDC117 | 0.658737 | 4 | 0 | 4 |
| OPA1 | SAAL1 | 0.656505 | 5 | 0 | 5 |
| OPA1 | SNAP47 | 0.654871 | 3 | 0 | 3 |
| OPA1 | ZNF367 | 0.651419 | 5 | 0 | 4 |
| OPA1 | MASTL | 0.649659 | 5 | 0 | 3 |
| OPA1 | PPIL1 | 0.6471 | 4 | 0 | 4 |
For details and further investigation, click here