| Full name: anaphase promoting complex subunit 13 | Alias Symbol: SWM1|APC13|DKFZP566D193 | ||
| Type: protein-coding gene | Cytoband: 3q22.2 | ||
| Entrez ID: 25847 | HGNC ID: HGNC:24540 | Ensembl Gene: ENSG00000129055 | OMIM ID: 614484 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ANAPC13 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation |
Expression of ANAPC13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ANAPC13 | 25847 | 209001_s_at | 0.1034 | 0.8264 | |
| GSE20347 | ANAPC13 | 25847 | 209001_s_at | 0.2215 | 0.2031 | |
| GSE23400 | ANAPC13 | 25847 | 209001_s_at | 0.4077 | 0.0000 | |
| GSE26886 | ANAPC13 | 25847 | 209001_s_at | 0.1941 | 0.3452 | |
| GSE29001 | ANAPC13 | 25847 | 209001_s_at | 0.4789 | 0.0950 | |
| GSE38129 | ANAPC13 | 25847 | 209001_s_at | 0.1141 | 0.3204 | |
| GSE45670 | ANAPC13 | 25847 | 209001_s_at | 0.1677 | 0.3578 | |
| GSE53622 | ANAPC13 | 25847 | 2321 | 0.4565 | 0.0000 | |
| GSE53624 | ANAPC13 | 25847 | 2321 | 0.5270 | 0.0000 | |
| GSE63941 | ANAPC13 | 25847 | 209001_s_at | -0.4068 | 0.2824 | |
| GSE77861 | ANAPC13 | 25847 | 209001_s_at | 0.2009 | 0.3523 | |
| GSE97050 | ANAPC13 | 25847 | A_32_P16854 | 0.3715 | 0.1955 | |
| SRP007169 | ANAPC13 | 25847 | RNAseq | 0.1767 | 0.7201 | |
| SRP008496 | ANAPC13 | 25847 | RNAseq | 0.4064 | 0.2093 | |
| SRP064894 | ANAPC13 | 25847 | RNAseq | 0.3033 | 0.1326 | |
| SRP133303 | ANAPC13 | 25847 | RNAseq | 0.4055 | 0.0014 | |
| SRP159526 | ANAPC13 | 25847 | RNAseq | 0.4932 | 0.1522 | |
| SRP193095 | ANAPC13 | 25847 | RNAseq | -0.0023 | 0.9880 | |
| SRP219564 | ANAPC13 | 25847 | RNAseq | 0.3431 | 0.3572 | |
| TCGA | ANAPC13 | 25847 | RNAseq | -0.0236 | 0.6612 |
Upregulated datasets: 0; Downregulated datasets: 0.
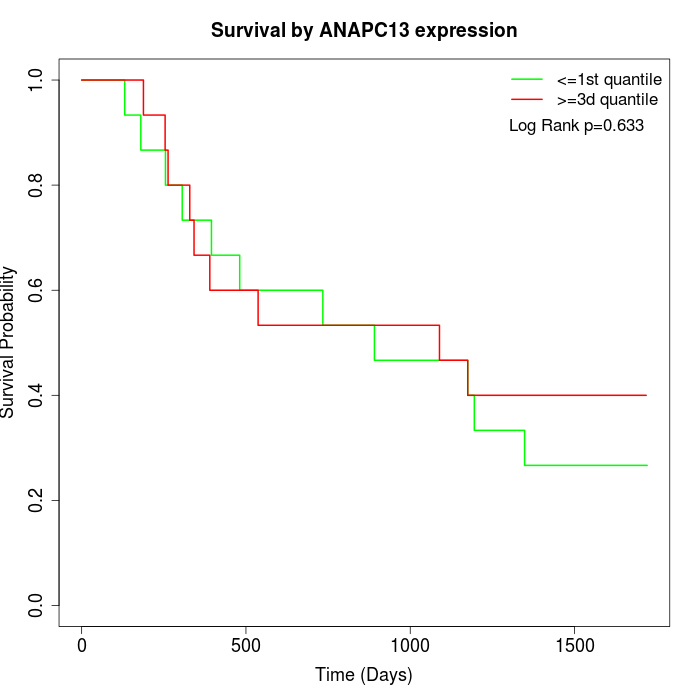
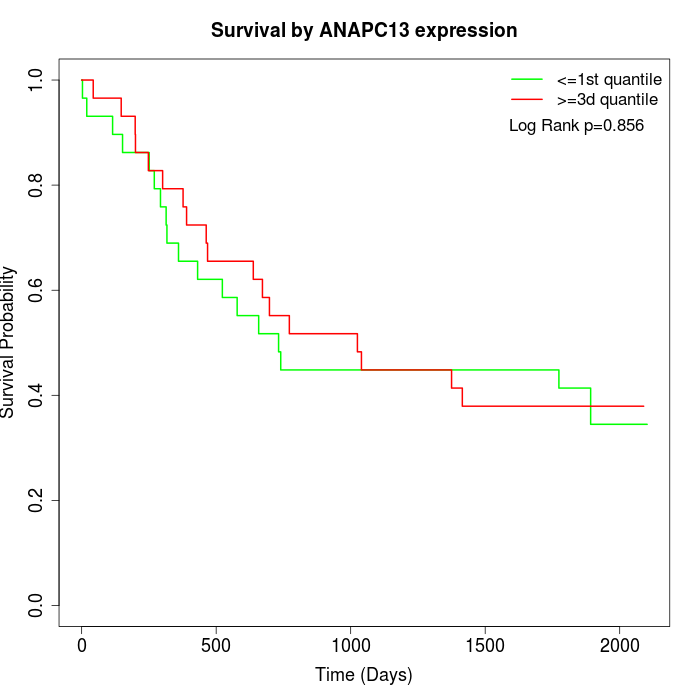
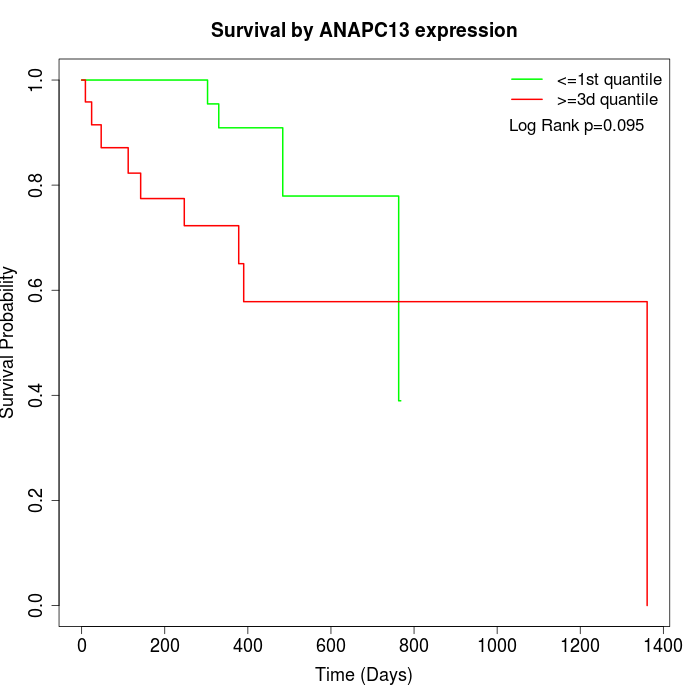
Survival by ANAPC13 expression:
Note: Click image to view full size file.
Copy number change of ANAPC13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ANAPC13 | 25847 | 16 | 0 | 14 | |
| GSE20123 | ANAPC13 | 25847 | 16 | 0 | 14 | |
| GSE43470 | ANAPC13 | 25847 | 21 | 0 | 22 | |
| GSE46452 | ANAPC13 | 25847 | 15 | 4 | 40 | |
| GSE47630 | ANAPC13 | 25847 | 18 | 3 | 19 | |
| GSE54993 | ANAPC13 | 25847 | 2 | 8 | 60 | |
| GSE54994 | ANAPC13 | 25847 | 32 | 1 | 20 | |
| GSE60625 | ANAPC13 | 25847 | 0 | 6 | 5 | |
| GSE74703 | ANAPC13 | 25847 | 18 | 0 | 18 | |
| GSE74704 | ANAPC13 | 25847 | 11 | 0 | 9 | |
| TCGA | ANAPC13 | 25847 | 59 | 5 | 32 |
Total number of gains: 208; Total number of losses: 27; Total Number of normals: 253.
Somatic mutations of ANAPC13:
Generating mutation plots.
Highly correlated genes for ANAPC13:
Showing top 20/597 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ANAPC13 | ZNF613 | 0.728068 | 3 | 0 | 3 |
| ANAPC13 | GTF2IRD1 | 0.727091 | 3 | 0 | 3 |
| ANAPC13 | YES1 | 0.726384 | 3 | 0 | 3 |
| ANAPC13 | PYCR2 | 0.725905 | 3 | 0 | 3 |
| ANAPC13 | CDC73 | 0.720471 | 3 | 0 | 3 |
| ANAPC13 | WDR54 | 0.715055 | 3 | 0 | 3 |
| ANAPC13 | ATG4C | 0.714208 | 3 | 0 | 3 |
| ANAPC13 | EIF4E2 | 0.706402 | 3 | 0 | 3 |
| ANAPC13 | ADO | 0.701123 | 4 | 0 | 4 |
| ANAPC13 | LARP1B | 0.700686 | 3 | 0 | 3 |
| ANAPC13 | EIF3M | 0.700368 | 3 | 0 | 3 |
| ANAPC13 | SUN1 | 0.699787 | 3 | 0 | 3 |
| ANAPC13 | XRN1 | 0.694093 | 3 | 0 | 3 |
| ANAPC13 | NUB1 | 0.693714 | 3 | 0 | 3 |
| ANAPC13 | FNIP2 | 0.693618 | 3 | 0 | 3 |
| ANAPC13 | SCO1 | 0.693255 | 3 | 0 | 3 |
| ANAPC13 | NF1 | 0.692256 | 3 | 0 | 3 |
| ANAPC13 | HSPA5 | 0.688815 | 3 | 0 | 3 |
| ANAPC13 | COX5B | 0.68299 | 3 | 0 | 3 |
| ANAPC13 | RBMX | 0.681778 | 3 | 0 | 3 |
For details and further investigation, click here