| Full name: acetyl-CoA carboxylase beta | Alias Symbol: HACC275|ACC2|ACCB | ||
| Type: protein-coding gene | Cytoband: 12q24.11 | ||
| Entrez ID: 32 | HGNC ID: HGNC:85 | Ensembl Gene: ENSG00000076555 | OMIM ID: 601557 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ACACB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04152 | AMPK signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04920 | Adipocytokine signaling pathway | |
| hsa04922 | Glucagon signaling pathway | |
| hsa04931 | Insulin resistance |
Expression of ACACB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACACB | 32 | 1552615_at | -0.2319 | 0.5077 | |
| GSE20347 | ACACB | 32 | 221928_at | -0.0671 | 0.6326 | |
| GSE23400 | ACACB | 32 | 221928_at | -0.6390 | 0.0000 | |
| GSE26886 | ACACB | 32 | 1552615_at | -0.2414 | 0.1594 | |
| GSE29001 | ACACB | 32 | 49452_at | -0.5213 | 0.4028 | |
| GSE38129 | ACACB | 32 | 221928_at | -0.8931 | 0.0012 | |
| GSE45670 | ACACB | 32 | 1552615_at | 0.0288 | 0.7523 | |
| GSE53622 | ACACB | 32 | 42642 | -1.6810 | 0.0000 | |
| GSE53624 | ACACB | 32 | 91464 | -0.9415 | 0.0000 | |
| GSE63941 | ACACB | 32 | 1552615_at | 0.3877 | 0.0287 | |
| GSE77861 | ACACB | 32 | 1552615_at | -0.0262 | 0.7423 | |
| GSE97050 | ACACB | 32 | A_33_P3334220 | -1.0949 | 0.1557 | |
| SRP007169 | ACACB | 32 | RNAseq | 0.9259 | 0.1412 | |
| SRP064894 | ACACB | 32 | RNAseq | -0.9079 | 0.0180 | |
| SRP133303 | ACACB | 32 | RNAseq | -1.1512 | 0.0037 | |
| SRP159526 | ACACB | 32 | RNAseq | -1.2282 | 0.0000 | |
| SRP193095 | ACACB | 32 | RNAseq | -0.5471 | 0.0401 | |
| SRP219564 | ACACB | 32 | RNAseq | -1.0086 | 0.1992 | |
| TCGA | ACACB | 32 | RNAseq | -1.1209 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
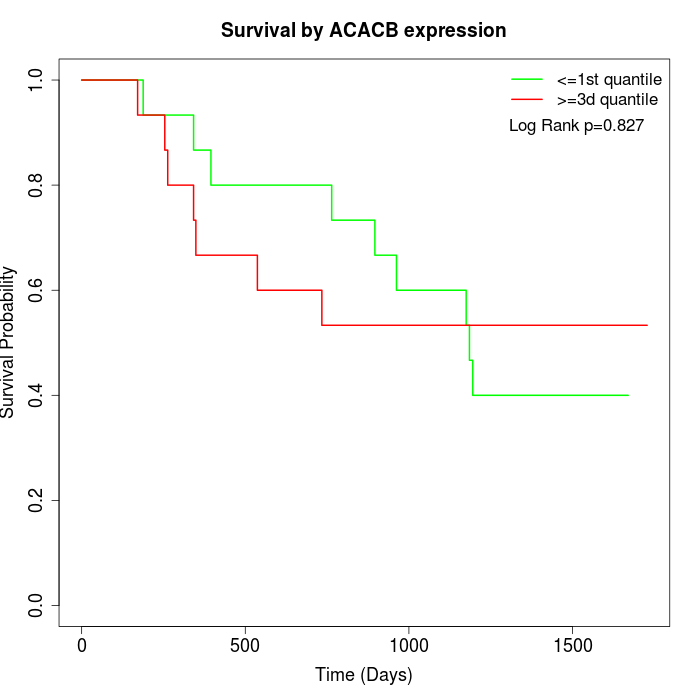
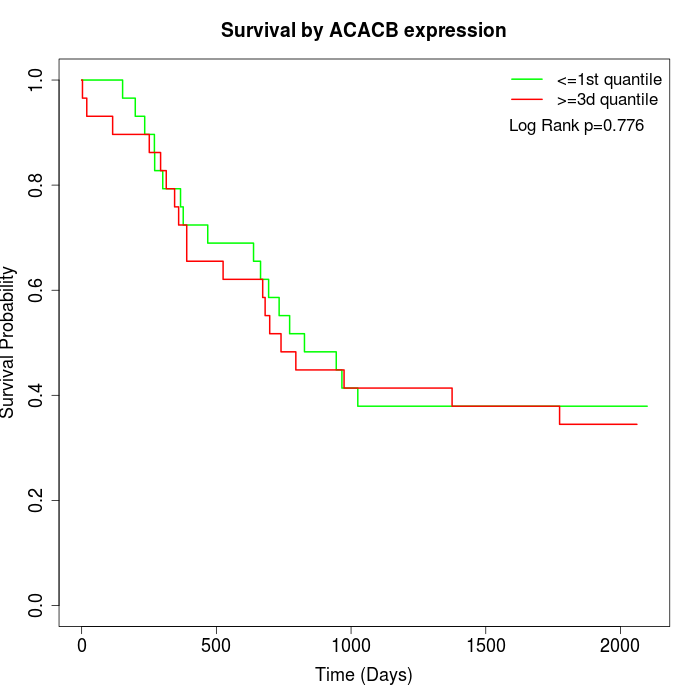
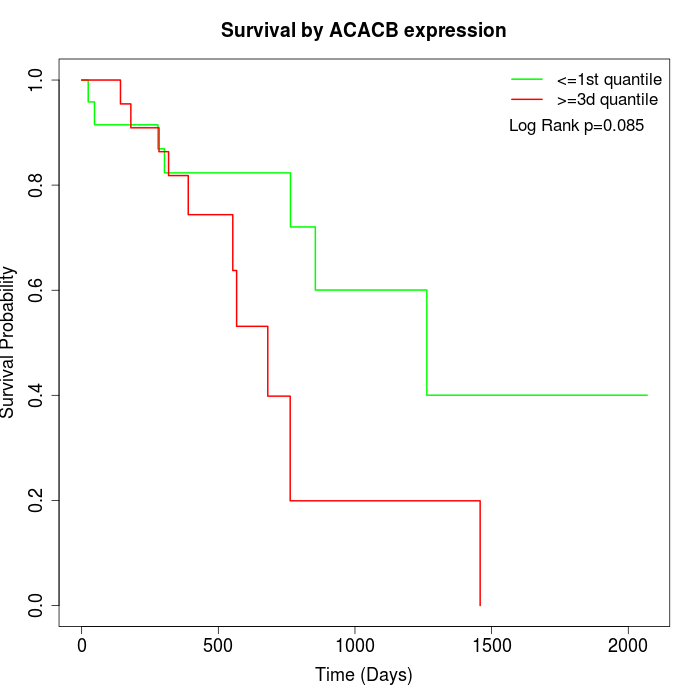
Survival by ACACB expression:
Note: Click image to view full size file.
Copy number change of ACACB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACACB | 32 | 5 | 3 | 22 | |
| GSE20123 | ACACB | 32 | 5 | 4 | 21 | |
| GSE43470 | ACACB | 32 | 2 | 1 | 40 | |
| GSE46452 | ACACB | 32 | 9 | 1 | 49 | |
| GSE47630 | ACACB | 32 | 9 | 1 | 30 | |
| GSE54993 | ACACB | 32 | 0 | 5 | 65 | |
| GSE54994 | ACACB | 32 | 4 | 3 | 46 | |
| GSE60625 | ACACB | 32 | 0 | 0 | 11 | |
| GSE74703 | ACACB | 32 | 2 | 0 | 34 | |
| GSE74704 | ACACB | 32 | 2 | 2 | 16 | |
| TCGA | ACACB | 32 | 21 | 10 | 65 |
Total number of gains: 59; Total number of losses: 30; Total Number of normals: 399.
Somatic mutations of ACACB:
Generating mutation plots.
Highly correlated genes for ACACB:
Showing top 20/621 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACACB | ARHGEF25 | 0.829748 | 3 | 0 | 3 |
| ACACB | PRR26 | 0.808705 | 3 | 0 | 3 |
| ACACB | ADAMTS9-AS2 | 0.774394 | 4 | 0 | 4 |
| ACACB | RYR2 | 0.752656 | 6 | 0 | 6 |
| ACACB | CADM2 | 0.742232 | 4 | 0 | 4 |
| ACACB | VMAC | 0.736517 | 3 | 0 | 3 |
| ACACB | MYOM1 | 0.735035 | 9 | 0 | 8 |
| ACACB | RBFOX3 | 0.729526 | 3 | 0 | 3 |
| ACACB | XKR4 | 0.727253 | 4 | 0 | 4 |
| ACACB | MITF | 0.725362 | 6 | 0 | 5 |
| ACACB | FXYD1 | 0.714115 | 7 | 0 | 6 |
| ACACB | CYP21A2 | 0.714095 | 3 | 0 | 3 |
| ACACB | ANGPTL7 | 0.705423 | 6 | 0 | 6 |
| ACACB | SMOC2 | 0.704074 | 4 | 0 | 4 |
| ACACB | PGR | 0.702663 | 5 | 0 | 4 |
| ACACB | LINC01119 | 0.702409 | 3 | 0 | 3 |
| ACACB | MEF2C-AS1 | 0.701402 | 3 | 0 | 3 |
| ACACB | RBPMS2 | 0.700352 | 5 | 0 | 4 |
| ACACB | ADCYAP1R1 | 0.699484 | 5 | 0 | 4 |
| ACACB | CBX7 | 0.699083 | 9 | 0 | 8 |
For details and further investigation, click here