| Full name: vimentin type intermediate filament associated coiled-coil protein | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 400673 | HGNC ID: HGNC:33803 | Ensembl Gene: ENSG00000187650 | OMIM ID: 617204 |
| Drug and gene relationship at DGIdb | |||
Expression of VMAC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | VMAC | 400673 | 69443 | -0.8222 | 0.0000 | |
| GSE53624 | VMAC | 400673 | 69443 | -0.8351 | 0.0000 | |
| GSE97050 | VMAC | 400673 | A_33_P3291176 | -1.1153 | 0.0818 | |
| SRP007169 | VMAC | 400673 | RNAseq | -2.2202 | 0.0000 | |
| SRP064894 | VMAC | 400673 | RNAseq | -0.4746 | 0.0092 | |
| SRP133303 | VMAC | 400673 | RNAseq | -0.5555 | 0.0004 | |
| SRP159526 | VMAC | 400673 | RNAseq | -0.4909 | 0.1227 | |
| SRP193095 | VMAC | 400673 | RNAseq | -0.4585 | 0.0010 | |
| SRP219564 | VMAC | 400673 | RNAseq | -0.4392 | 0.3041 | |
| TCGA | VMAC | 400673 | RNAseq | -0.1285 | 0.1200 |
Upregulated datasets: 0; Downregulated datasets: 1.
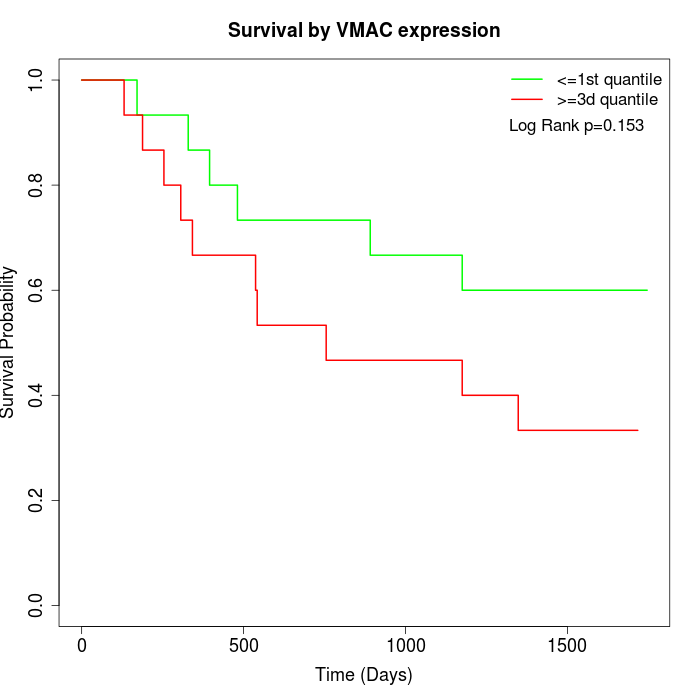
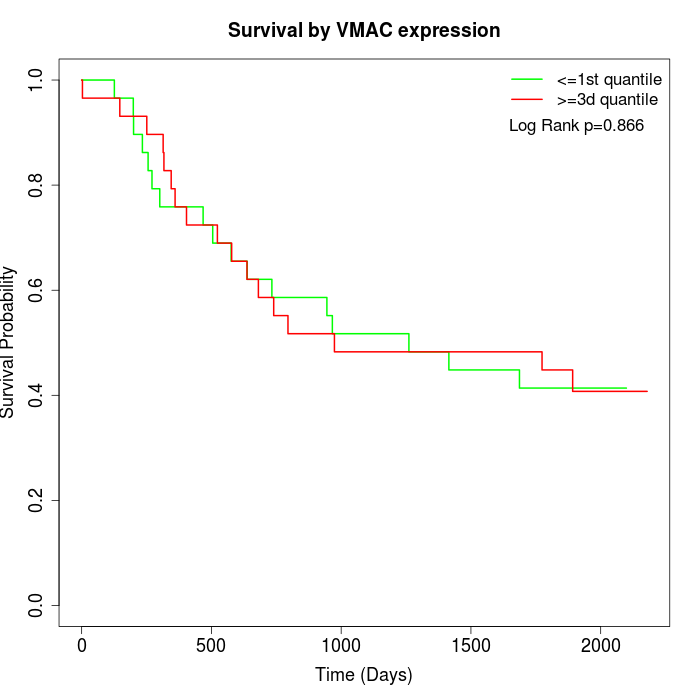
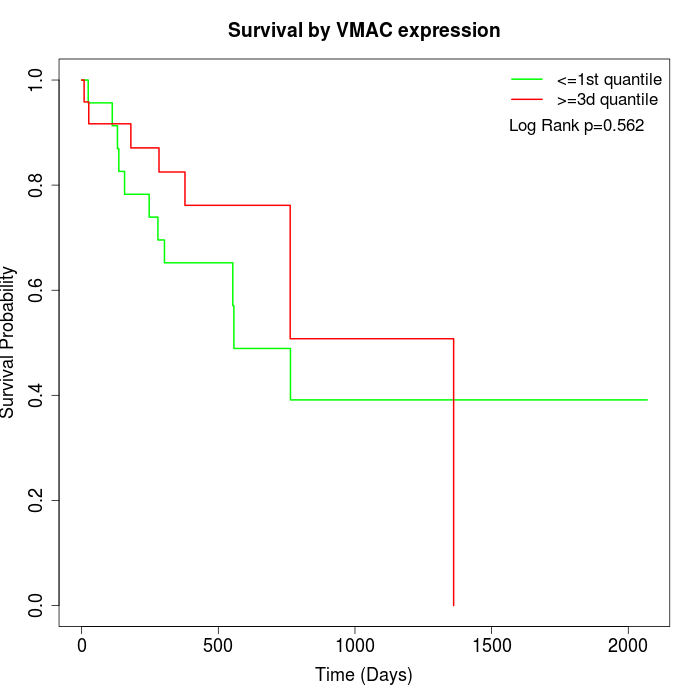
Survival by VMAC expression:
Note: Click image to view full size file.
Copy number change of VMAC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VMAC | 400673 | 5 | 3 | 22 | |
| GSE20123 | VMAC | 400673 | 4 | 2 | 24 | |
| GSE43470 | VMAC | 400673 | 2 | 6 | 35 | |
| GSE46452 | VMAC | 400673 | 47 | 1 | 11 | |
| GSE47630 | VMAC | 400673 | 5 | 7 | 28 | |
| GSE54993 | VMAC | 400673 | 16 | 3 | 51 | |
| GSE54994 | VMAC | 400673 | 6 | 16 | 31 | |
| GSE60625 | VMAC | 400673 | 9 | 0 | 2 | |
| GSE74703 | VMAC | 400673 | 2 | 4 | 30 | |
| GSE74704 | VMAC | 400673 | 1 | 2 | 17 | |
| TCGA | VMAC | 400673 | 9 | 18 | 69 |
Total number of gains: 106; Total number of losses: 62; Total Number of normals: 320.
Somatic mutations of VMAC:
Generating mutation plots.
Highly correlated genes for VMAC:
Showing top 20/176 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VMAC | MYOCD | 0.851075 | 3 | 0 | 3 |
| VMAC | HMHB1 | 0.82195 | 3 | 0 | 3 |
| VMAC | ANKS1B | 0.807383 | 3 | 0 | 3 |
| VMAC | SLC2A4 | 0.800252 | 3 | 0 | 3 |
| VMAC | LMOD1 | 0.797486 | 3 | 0 | 3 |
| VMAC | PYGM | 0.797442 | 3 | 0 | 3 |
| VMAC | MYL5 | 0.792753 | 3 | 0 | 3 |
| VMAC | CACNB2 | 0.789407 | 3 | 0 | 3 |
| VMAC | DES | 0.787526 | 3 | 0 | 3 |
| VMAC | LIMS2 | 0.786611 | 3 | 0 | 3 |
| VMAC | SYNM | 0.784775 | 3 | 0 | 3 |
| VMAC | HPSE2 | 0.780701 | 3 | 0 | 3 |
| VMAC | INMT | 0.777762 | 3 | 0 | 3 |
| VMAC | PCP4 | 0.775542 | 3 | 0 | 3 |
| VMAC | PDE1C | 0.77245 | 3 | 0 | 3 |
| VMAC | PPP1R12B | 0.771006 | 3 | 0 | 3 |
| VMAC | PLP1 | 0.76959 | 3 | 0 | 3 |
| VMAC | FLNC | 0.768733 | 3 | 0 | 3 |
| VMAC | ATP1A2 | 0.768698 | 3 | 0 | 3 |
| VMAC | SORBS1 | 0.766965 | 3 | 0 | 3 |
For details and further investigation, click here